Molecolab Pisa
@molecolabpisa.bsky.social
88 followers
88 following
10 posts
Computational Chemistry Research Group at the University of Pisa, Italy https://molecolab.dcci.unipi.it/
Posts
Media
Videos
Starter Packs
Molecolab Pisa
@molecolabpisa.bsky.social
· Aug 29

An Efficient and Robust Implementation of CASSCF Linear Response Theory
We present a robust and efficient implementation of linear response theory for a Complete Active Space─Self-Consistent Field wave function. Our approach relies on the Cholesky Decomposition of the two-electron integrals, enabling the routine treatment of large molecular systems on standard hardware. It allows for the computation of both absorption energies and transition properties, as well as frequency-dependent molecular response functions. For both classes of properties, numerically stable and efficient algorithms have been developed. The capabilities of our implementation are demonstrated through the calculation of absorption spectra and molecular response properties of large systems with extended basis sets.
doi.org
Molecolab Pisa
@molecolabpisa.bsky.social
· Aug 28

Multiscale Simulation of Photoinduced Electron Transfer in Cryptochrome 4 from European Robin and Pigeon Indicates a Conserved Dynamics
Cryptochrome 4 (Cry4) is a leading candidate for mediating magnetoreception in birds. Upon photoexcitation of its flavin adenine dinucleotide (FAD) cofactor, Cry4 initiates an electron transfer (ET) c...
doi.org
Reposted by Molecolab Pisa
Molecolab Pisa
@molecolabpisa.bsky.social
· Jan 20

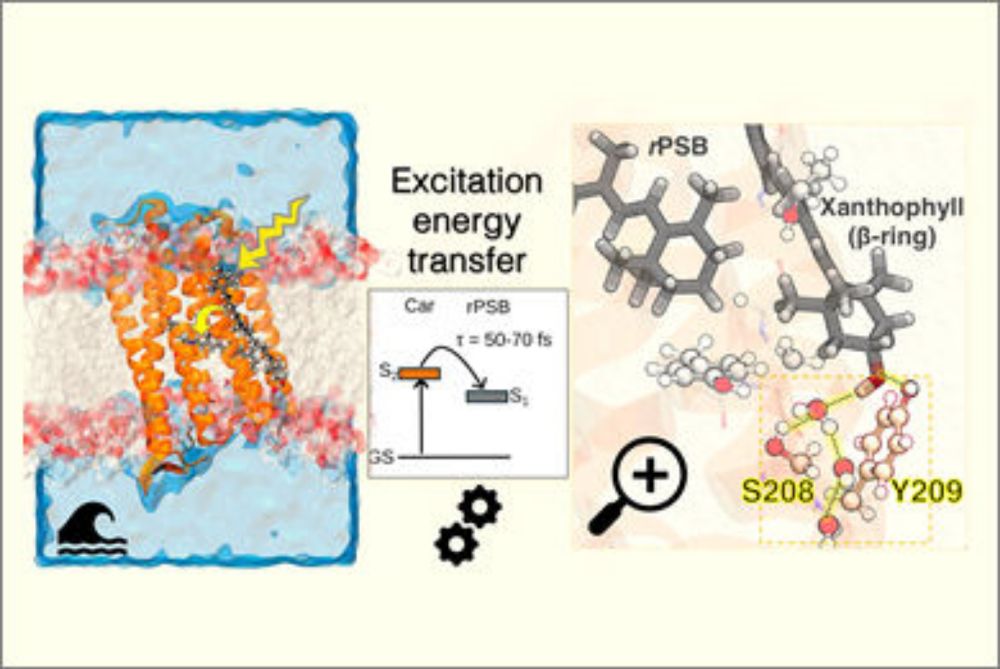
A Computational Approach to Modeling Excitation Energy Transfer and Quenching in Light-Harvesting Complexes
Light-harvesting complexes (LHCs) play a critical role in modulating energy flux within photosynthetic organisms in response to fluctuating light. Under high light conditions, they activate quenching ...
doi.org
Molecolab Pisa
@molecolabpisa.bsky.social
· Dec 20

Protein-Driven Electron-Transfer Process in a Fatty Acid Photodecarboxylase
Naturally occurring photoenzymes are rare in nature, but among them, fatty acid photodecarboxylases derived from Chlorella variabilis (CvFAPs) have emerged as promising photobiocatalysts capable of performing the redox-neutral, light-induced decarboxylation of free fatty acids (FAs) into C1-shortened n-alka(e)nes. Using a hybrid QM/MM approach combined with a polarizable embedding scheme, we identify the structural changes of the active site and determine the energetic landscape of the forward electron transfer (fET) from the FA substrate to the excited flavin adenine dinucleotide. We obtain a charge-transfer diradical structure where a water molecule rearranges spontaneously to form a H-bond interaction with the excited flavin, while the FA’s carboxylate group twists and migrates away from it. Together, these structural modifications provide the driving force necessary for the fET to proceed in a downhill direction. Moreover, by examining the R451K mutant where the FA substrate is farther from the flavin core, we show that the marked reduction of the electronic coupling is counterbalanced by an increased driving force, resulting in a fET lifetime similar to the WT, thereby suggesting a resilience of the process to this mutation. Finally, through QM/MM molecular dynamic simulations, we reveal that, following fET, the decarboxylation of the FA radical occurs within tens of picoseconds, overcoming an energy barrier of ∼0.1 eV. Overall, by providing an atomistic characterization of the photoactivation of CvFAP, this work can be used for future protein engineering.
pubs.acs.org