Petr Šulc
@petrsulc.bsky.social
580 followers
910 following
34 posts
Associate Professor at Arizona State University, and ERC grant PI at TU Munich. We play with DNA and RNA to make nanoscale structures and devices. Find out more at our lab page: sulclab.org
Posts
Media
Videos
Starter Packs
Reposted by Petr Šulc
Reposted by Petr Šulc
Petr Šulc
@petrsulc.bsky.social
· Jun 7
Petr Šulc
@petrsulc.bsky.social
· Jun 7
Petr Šulc
@petrsulc.bsky.social
· Jun 7
Petr Šulc
@petrsulc.bsky.social
· Apr 26
Petr Šulc
@petrsulc.bsky.social
· Apr 8
Petr Šulc
@petrsulc.bsky.social
· Apr 8
Petr Šulc
@petrsulc.bsky.social
· Apr 8
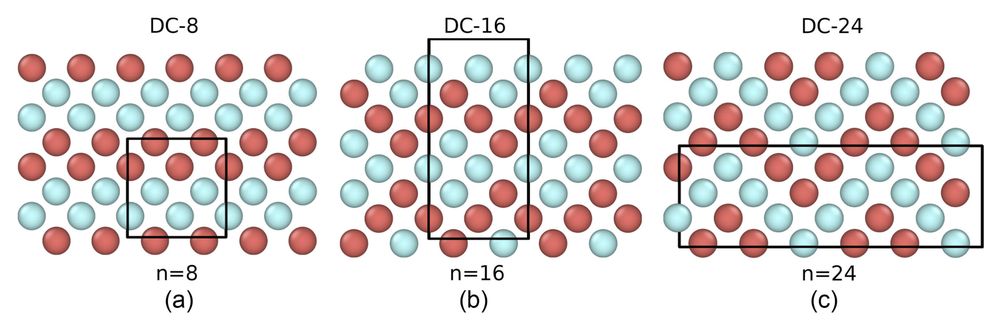
Falsifiability Test for Classical Nucleation Theory
Classical nucleation theory (CNT) is built upon the capillarity approximation, i.e., the assumption that the nucleation properties can be inferred from the bulk properties of the melt and the crystal....
journals.aps.org
Petr Šulc
@petrsulc.bsky.social
· Mar 24
Petr Šulc
@petrsulc.bsky.social
· Mar 17
Aidin Lak
@laklab-tubs.bsky.social
· Mar 17

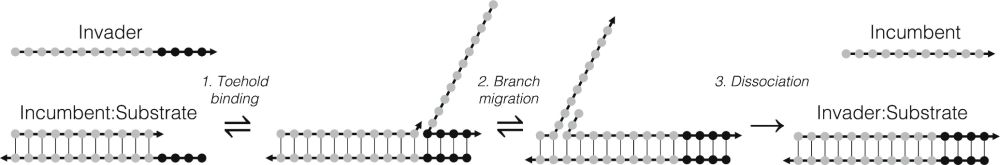
Mismatch-Assisted Toehold Exchange Cascades for Magnetic Nanoparticle-Based Nucleic Acid Diagnostics
Sensitive, simple, and rapid detection of nucleic acid sequences at point-of-care (POC) settings is still an unmet quest. Magnetic readout assays combined with toehold-mediated strand displacement (TM...
biorxiv.org
Petr Šulc
@petrsulc.bsky.social
· Feb 5
Petr Šulc
@petrsulc.bsky.social
· Jan 20