Sara Carioscia
@saracarioscia.bsky.social
160 followers
390 following
22 posts
Human genetics & evolution | PhD student with Rajiv McCoy at Johns Hopkins | Bikes | https://scarioscia.github.io/
Posts
Media
Videos
Starter Packs
Reposted by Sara Carioscia
Matthew Aguirre
@aguirre404.bsky.social
· Aug 22
Reposted by Sara Carioscia
Reposted by Sara Carioscia
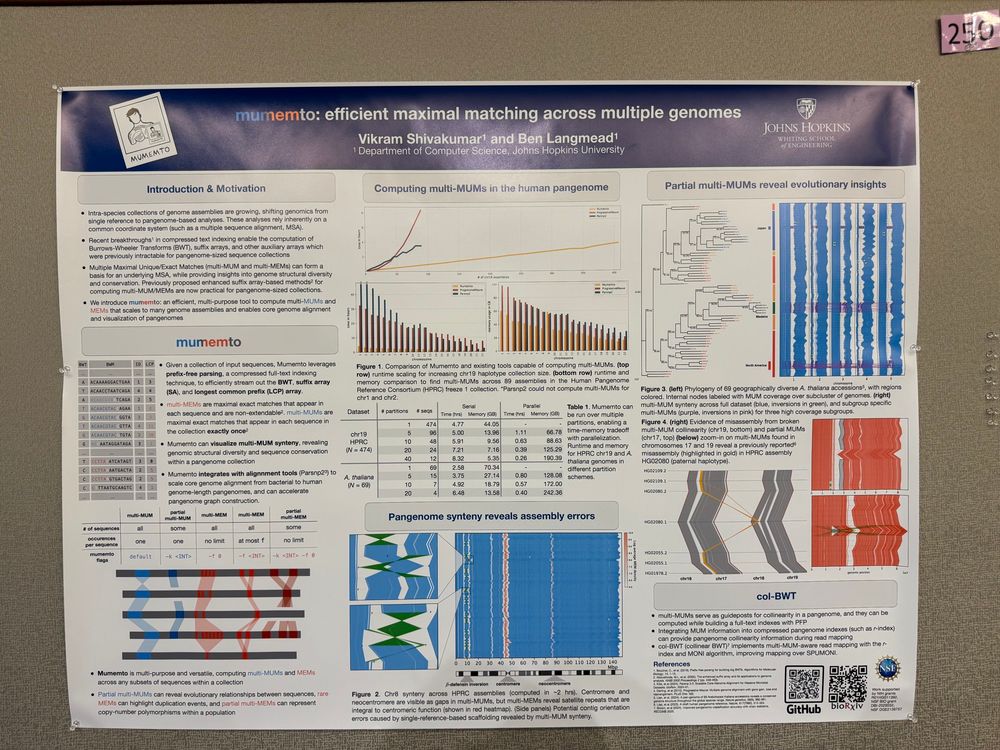
Ben Langmead
@benlangmead.bsky.social
· Jun 17
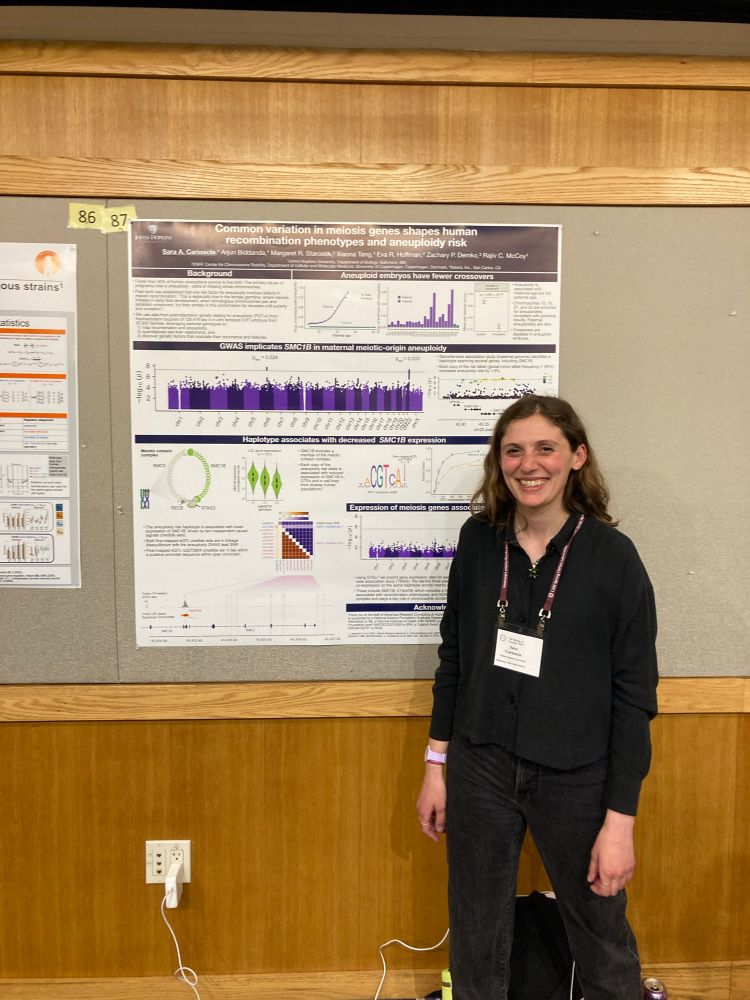
Sara Carioscia
@saracarioscia.bsky.social
· May 16
Reposted by Sara Carioscia
Arjun Biddanda
@aabiddanda.github.io
· May 15
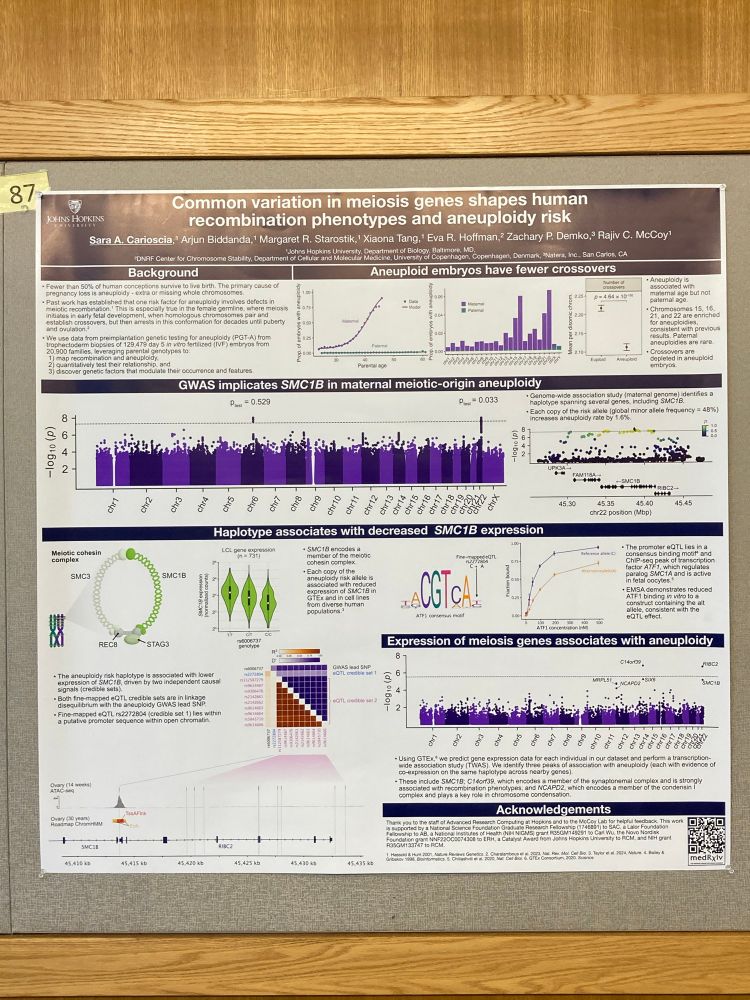
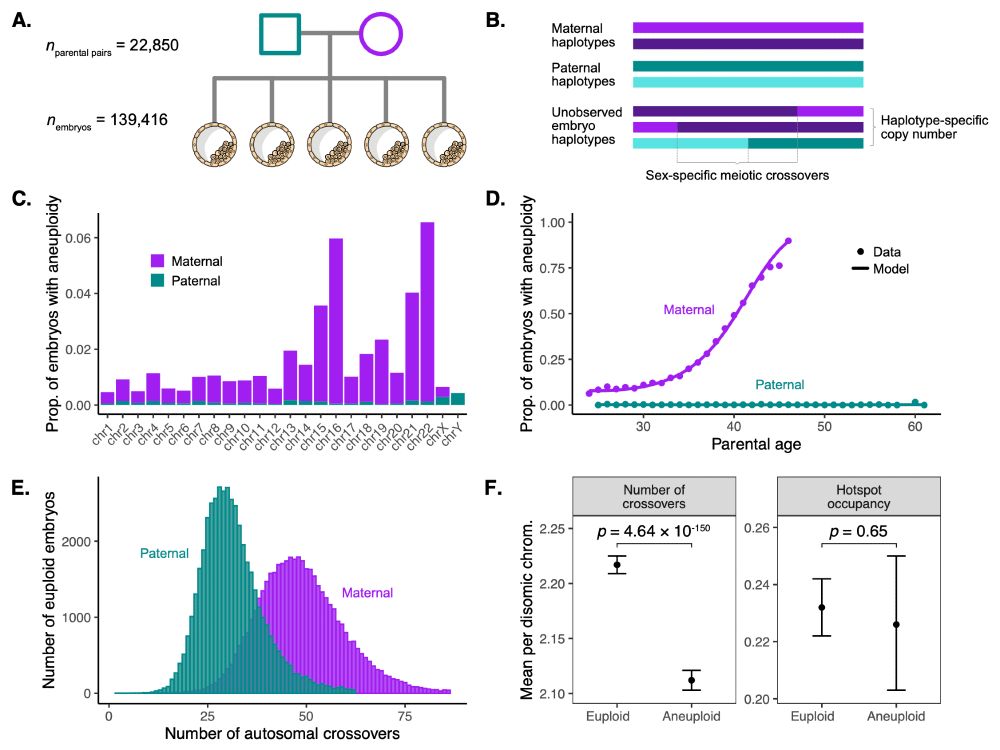
Sara Carioscia
@saracarioscia.bsky.social
· May 15
Reposted by Sara Carioscia
Sara Carioscia
@saracarioscia.bsky.social
· Apr 28
Sara Carioscia
@saracarioscia.bsky.social
· Apr 26
Sara Carioscia
@saracarioscia.bsky.social
· Apr 24
Sara Carioscia
@saracarioscia.bsky.social
· Apr 24
Reposted by Sara Carioscia
Arjun Biddanda
@aabiddanda.github.io
· Apr 7
Reposted by Sara Carioscia