Engin Yapici
@enginyapici.bsky.social
Drug discovery scientist writing about bioassays, AI in biotech, and the messy, fascinating process of turning biology into medicine.
Pinned
Engin Yapici
@enginyapici.bsky.social
· Apr 13
Hi, I’m Engin. I work in drug discovery, mostly antibody discovery and development, high-throughput screening, and MoA functional assays. I’m here to share what I’ve learned, what I’m still figuring out, and to learn from others thinking deeply about how we move this field forward.
New plate-based HIC assay: 96-well, ~50 µg per sample, full readout in 2 hrs. Better dynamic range than AC-SINS, closer to true aHIC. Flags high-risk antibodies early without the bottleneck.
medium.com/@enginyapici...
medium.com/@enginyapici...

A High‑Throughput HIC Assay for Antibody Developability
A plate-based surrogate HIC assay cuts screening time from days to hours with only 50 µg of antibody per sample
medium.com
October 1, 2025 at 5:11 AM
New plate-based HIC assay: 96-well, ~50 µg per sample, full readout in 2 hrs. Better dynamic range than AC-SINS, closer to true aHIC. Flags high-risk antibodies early without the bottleneck.
medium.com/@enginyapici...
medium.com/@enginyapici...
Just published: 𝗪𝗵𝗮𝘁 𝗩𝗶𝗿𝘁𝘂𝗮𝗹 𝗖𝗲𝗹𝗹𝘀 𝗦𝘁𝗶𝗹𝗹 𝗡𝗲𝗲𝗱
medium.com/@enginyapici...
Recursion and Valence outline a big vision for modeling biology. This post adds what I think are still-missing layers: metabolite-driven regulation, protein-level function, and failure-based learning.
#Biotech #AIHealthcare
medium.com/@enginyapici...
Recursion and Valence outline a big vision for modeling biology. This post adds what I think are still-missing layers: metabolite-driven regulation, protein-level function, and failure-based learning.
#Biotech #AIHealthcare

What Virtual Cells Still Need: Protein-Level Function, Metabolites, and Beyond
A complementary perspective on Recursion and Valence Labs’ recent “Virtual Cells” roadmap
medium.com
June 16, 2025 at 6:30 PM
Just published: 𝗪𝗵𝗮𝘁 𝗩𝗶𝗿𝘁𝘂𝗮𝗹 𝗖𝗲𝗹𝗹𝘀 𝗦𝘁𝗶𝗹𝗹 𝗡𝗲𝗲𝗱
medium.com/@enginyapici...
Recursion and Valence outline a big vision for modeling biology. This post adds what I think are still-missing layers: metabolite-driven regulation, protein-level function, and failure-based learning.
#Biotech #AIHealthcare
medium.com/@enginyapici...
Recursion and Valence outline a big vision for modeling biology. This post adds what I think are still-missing layers: metabolite-driven regulation, protein-level function, and failure-based learning.
#Biotech #AIHealthcare
Just wrote about a platform that pulled out 𝟱 𝘀𝘂𝗯-𝗻𝗮𝗻𝗼𝗺𝗼𝗹𝗮𝗿 antibodies in 3 weeks. From plasma cells, not display libraries.
Naturally paired, functionally diverse, and validated early (blockers, agonists, bins).
medium.com/@enginyapici...
#AntibodyDiscovery #DrugDiscovery #Microfluidics #Biotech
Naturally paired, functionally diverse, and validated early (blockers, agonists, bins).
medium.com/@enginyapici...
#AntibodyDiscovery #DrugDiscovery #Microfluidics #Biotech

Why the Next Great Antibody May Come from a Droplet, Not a Mouse Spleen
How a droplet-based system found sub-nanomolar antibodies in weeks, not months
medium.com
June 13, 2025 at 7:36 AM
Just wrote about a platform that pulled out 𝟱 𝘀𝘂𝗯-𝗻𝗮𝗻𝗼𝗺𝗼𝗹𝗮𝗿 antibodies in 3 weeks. From plasma cells, not display libraries.
Naturally paired, functionally diverse, and validated early (blockers, agonists, bins).
medium.com/@enginyapici...
#AntibodyDiscovery #DrugDiscovery #Microfluidics #Biotech
Naturally paired, functionally diverse, and validated early (blockers, agonists, bins).
medium.com/@enginyapici...
#AntibodyDiscovery #DrugDiscovery #Microfluidics #Biotech
65 designs. Single shot. 16 recovered binding to XBB.1.5.
No iterative wet-lab cycles. No massive screens.
They solve three big problems in one shot: escape recovery, developability, diversity.
I broke it down here:
medium.com/@enginyapici...
#AntibodyEngieering #Biotech #AntibodyDiscovery #AI
No iterative wet-lab cycles. No massive screens.
They solve three big problems in one shot: escape recovery, developability, diversity.
I broke it down here:
medium.com/@enginyapici...
#AntibodyEngieering #Biotech #AntibodyDiscovery #AI

Doing It All: A Rare Triple Play in Computational Antibody Design
How one team tackled escape mutations, developability, and diversity, with just 65 designs
medium.com
June 10, 2025 at 6:07 AM
65 designs. Single shot. 16 recovered binding to XBB.1.5.
No iterative wet-lab cycles. No massive screens.
They solve three big problems in one shot: escape recovery, developability, diversity.
I broke it down here:
medium.com/@enginyapici...
#AntibodyEngieering #Biotech #AntibodyDiscovery #AI
No iterative wet-lab cycles. No massive screens.
They solve three big problems in one shot: escape recovery, developability, diversity.
I broke it down here:
medium.com/@enginyapici...
#AntibodyEngieering #Biotech #AntibodyDiscovery #AI
What if your assay failed because two proteins shared a ligand you didn’t track?
Entabolons = proteins functionally linked by the same metabolite. No interaction, no pathway step: just a shared dependency missing from most models.
medium.com/@enginyapici...
#drugdiscovery #systemsbiology
Entabolons = proteins functionally linked by the same metabolite. No interaction, no pathway step: just a shared dependency missing from most models.
medium.com/@enginyapici...
#drugdiscovery #systemsbiology
Entabolons Are Just the Missing Layer Between Metabolites and Interactomes
How understanding entabolons can help us build better drug discovery assays
medium.com
May 30, 2025 at 5:16 AM
What if your assay failed because two proteins shared a ligand you didn’t track?
Entabolons = proteins functionally linked by the same metabolite. No interaction, no pathway step: just a shared dependency missing from most models.
medium.com/@enginyapici...
#drugdiscovery #systemsbiology
Entabolons = proteins functionally linked by the same metabolite. No interaction, no pathway step: just a shared dependency missing from most models.
medium.com/@enginyapici...
#drugdiscovery #systemsbiology
The FDA’s draft AI guidance treats assistive tools like decision-makers. That’s a problem. Most AI helps teams triage or prioritize, not drive filings.
Here’s my take on how this could backfire for biotech teams or become an edge for first-time filers: medium.com/@enginyapici...
#biotech #fda #ai
Here’s my take on how this could backfire for biotech teams or become an edge for first-time filers: medium.com/@enginyapici...
#biotech #fda #ai
When AI Transparency Becomes Bureaucracy: Reflections on the FDA’s New Draft Guidance
The FDA’s new draft guidance, Considerations for the Use of Artificial Intelligence To Support Regulatory Decision-Making for Drug and…
medium.com
May 25, 2025 at 7:40 PM
The FDA’s draft AI guidance treats assistive tools like decision-makers. That’s a problem. Most AI helps teams triage or prioritize, not drive filings.
Here’s my take on how this could backfire for biotech teams or become an edge for first-time filers: medium.com/@enginyapici...
#biotech #fda #ai
Here’s my take on how this could backfire for biotech teams or become an edge for first-time filers: medium.com/@enginyapici...
#biotech #fda #ai
Most AI antibody papers talk models. This one talks infrastructure.
Ginkgo’s PROPHET-Ab platform runs real assays, at scale, upstream, and cleanly. But can it handle messy, early-stage variants?
medium.com/@enginyapici...
#DrugDevelopment #AIinBiotech #Antibodies #DrugDiscovery #Biologics #AI
Ginkgo’s PROPHET-Ab platform runs real assays, at scale, upstream, and cleanly. But can it handle messy, early-stage variants?
medium.com/@enginyapici...
#DrugDevelopment #AIinBiotech #Antibodies #DrugDiscovery #Biologics #AI

What It Looks Like to Industrialize Antibody Developability Assays
A proof of what becomes possible when assay platforms are built with AI-scale data generation in mind.
medium.com
May 20, 2025 at 4:48 AM
Most AI antibody papers talk models. This one talks infrastructure.
Ginkgo’s PROPHET-Ab platform runs real assays, at scale, upstream, and cleanly. But can it handle messy, early-stage variants?
medium.com/@enginyapici...
#DrugDevelopment #AIinBiotech #Antibodies #DrugDiscovery #Biologics #AI
Ginkgo’s PROPHET-Ab platform runs real assays, at scale, upstream, and cleanly. But can it handle messy, early-stage variants?
medium.com/@enginyapici...
#DrugDevelopment #AIinBiotech #Antibodies #DrugDiscovery #Biologics #AI
𝗬𝗼𝘂 𝗰𝗮𝗻 𝗻𝗼𝘄 𝘄𝗮𝘁𝗰𝗵 𝗺𝗼𝗹𝗲𝗰𝘂𝗹𝗲𝘀 𝗰𝗵𝗮𝗻𝗴𝗲 𝘀𝗵𝗮𝗽𝗲 𝗶𝗻 𝗿𝗲𝗮𝗹 𝘁𝗶𝗺𝗲.
It measures how long a single molecule stays trapped, and turns that into size, shape, and binding data. No freezing, no tethering, no guessing.
medium.com/@enginyapici...
#Biotech #DrugDiscovery #ProteinStructure #StructuralBiology
It measures how long a single molecule stays trapped, and turns that into size, shape, and binding data. No freezing, no tethering, no guessing.
medium.com/@enginyapici...
#Biotech #DrugDiscovery #ProteinStructure #StructuralBiology

You Can Now Watch Molecules Change Shape in Real Time
We finally have a way to measure structural change in solution, in real time, without freezing, tethering, or guessing.
medium.com
May 12, 2025 at 6:17 PM
𝗬𝗼𝘂 𝗰𝗮𝗻 𝗻𝗼𝘄 𝘄𝗮𝘁𝗰𝗵 𝗺𝗼𝗹𝗲𝗰𝘂𝗹𝗲𝘀 𝗰𝗵𝗮𝗻𝗴𝗲 𝘀𝗵𝗮𝗽𝗲 𝗶𝗻 𝗿𝗲𝗮𝗹 𝘁𝗶𝗺𝗲.
It measures how long a single molecule stays trapped, and turns that into size, shape, and binding data. No freezing, no tethering, no guessing.
medium.com/@enginyapici...
#Biotech #DrugDiscovery #ProteinStructure #StructuralBiology
It measures how long a single molecule stays trapped, and turns that into size, shape, and binding data. No freezing, no tethering, no guessing.
medium.com/@enginyapici...
#Biotech #DrugDiscovery #ProteinStructure #StructuralBiology
Just published a new piece: what real wet-lab validation should look like in AI-enabled antibody design.
I walk through what’s often missing: scaffold diversity, expression, off-target data, developability. And why these matter if we want the models to translate.
medium.com/@enginyapici...
I walk through what’s often missing: scaffold diversity, expression, off-target data, developability. And why these matter if we want the models to translate.
medium.com/@enginyapici...

A Guide to Real Validation in AI-Enabled Antibody Design
Think your model worked? Then show me the gating strategy, binding curves, sensorgrams, and yields. Otherwise, you’ve got a sequence, not a…
medium.com
May 5, 2025 at 3:20 PM
Just published a new piece: what real wet-lab validation should look like in AI-enabled antibody design.
I walk through what’s often missing: scaffold diversity, expression, off-target data, developability. And why these matter if we want the models to translate.
medium.com/@enginyapici...
I walk through what’s often missing: scaffold diversity, expression, off-target data, developability. And why these matter if we want the models to translate.
medium.com/@enginyapici...
Can AI agents really design complex scientific workflows?
I wrote about a new benchmark that puts autonomous systems to the test: no handholding, no domain hints.
Where they shine, where they fail, and what it means for drug discovery.
medium.com/@enginyapici...
#DrugDiscovery #AIDrugDiscovery
I wrote about a new benchmark that puts autonomous systems to the test: no handholding, no domain hints.
Where they shine, where they fail, and what it means for drug discovery.
medium.com/@enginyapici...
#DrugDiscovery #AIDrugDiscovery

How Close Are We to AI Agents Designing Complex Scientific Workflows?
What a recent drug discovery benchmark reveals about the limits of autonomous AI agents
medium.com
May 1, 2025 at 6:25 AM
Can AI agents really design complex scientific workflows?
I wrote about a new benchmark that puts autonomous systems to the test: no handholding, no domain hints.
Where they shine, where they fail, and what it means for drug discovery.
medium.com/@enginyapici...
#DrugDiscovery #AIDrugDiscovery
I wrote about a new benchmark that puts autonomous systems to the test: no handholding, no domain hints.
Where they shine, where they fail, and what it means for drug discovery.
medium.com/@enginyapici...
#DrugDiscovery #AIDrugDiscovery
Just published a new piece:
Building Better Antibodies: Lessons from SynAbLib and IgHuAb
How large language models are helping design human-like antibody libraries that are actually usable for discovery.
medium.com/@enginyapici...
#AntibodyDiscovery #Biotechnology #MachineLearning #DrugDiscovery
Building Better Antibodies: Lessons from SynAbLib and IgHuAb
How large language models are helping design human-like antibody libraries that are actually usable for discovery.
medium.com/@enginyapici...
#AntibodyDiscovery #Biotechnology #MachineLearning #DrugDiscovery

Building Better Antibodies: Lessons from SynAbLib and IgHuAb
Using large language models to build scalable, human-like synthetic antibody libraries for therapeutic discovery and antibody engineering.
medium.com
April 27, 2025 at 8:37 PM
Just published a new piece:
Building Better Antibodies: Lessons from SynAbLib and IgHuAb
How large language models are helping design human-like antibody libraries that are actually usable for discovery.
medium.com/@enginyapici...
#AntibodyDiscovery #Biotechnology #MachineLearning #DrugDiscovery
Building Better Antibodies: Lessons from SynAbLib and IgHuAb
How large language models are helping design human-like antibody libraries that are actually usable for discovery.
medium.com/@enginyapici...
#AntibodyDiscovery #Biotechnology #MachineLearning #DrugDiscovery
This post explores the gap between AI tools in drug discovery and the scientists who need them. I highlight a smart low-data model and share thoughts on how better collaboration could make it truly usable.
www.linkedin.com/pulse/ai-dru...
#GenerativeAI #DrugDiscovery #AIDrugDiscovery #AI #ML
www.linkedin.com/pulse/ai-dru...
#GenerativeAI #DrugDiscovery #AIDrugDiscovery #AI #ML

In AI Drug Discovery, the Problem Isn’t the Model: It’s the Handoff
There’s no shortage of innovation in computational drug discovery right now. Every month, we see new models, better benchmarks, and smarter architectures.
www.linkedin.com
April 21, 2025 at 5:13 PM
This post explores the gap between AI tools in drug discovery and the scientists who need them. I highlight a smart low-data model and share thoughts on how better collaboration could make it truly usable.
www.linkedin.com/pulse/ai-dru...
#GenerativeAI #DrugDiscovery #AIDrugDiscovery #AI #ML
www.linkedin.com/pulse/ai-dru...
#GenerativeAI #DrugDiscovery #AIDrugDiscovery #AI #ML
Can AI predict high-viscosity mAbs without a structure?
DeepViscosity uses antibody sequence alone to flag formulation risks, before any wet-lab work. I break down what the model does well, where it fits in real workflows, and how it compares to other tools.
medium.com/@enginyapici...
#biologics
DeepViscosity uses antibody sequence alone to flag formulation risks, before any wet-lab work. I break down what the model does well, where it fits in real workflows, and how it compares to other tools.
medium.com/@enginyapici...
#biologics

Can We Predict High-Viscosity mAbs Without a Structure?
A look at DeepViscosity and how ensemble learning could save time and material in high-concentration antibody formulation
medium.com
April 21, 2025 at 12:29 AM
Can AI predict high-viscosity mAbs without a structure?
DeepViscosity uses antibody sequence alone to flag formulation risks, before any wet-lab work. I break down what the model does well, where it fits in real workflows, and how it compares to other tools.
medium.com/@enginyapici...
#biologics
DeepViscosity uses antibody sequence alone to flag formulation risks, before any wet-lab work. I break down what the model does well, where it fits in real workflows, and how it compares to other tools.
medium.com/@enginyapici...
#biologics
Can generative AI design antibodies without ever stepping into a lab?
I wrote about PG-AbD, a solid framework with no wet-lab validation, and why that's not a dead end, just a missed opportunity.
medium.com/@enginyapici...
#AI #DrugDiscovery #AntibodyDiscovery #AIinBiotech #Biotech #GenerativeAI
I wrote about PG-AbD, a solid framework with no wet-lab validation, and why that's not a dead end, just a missed opportunity.
medium.com/@enginyapici...
#AI #DrugDiscovery #AntibodyDiscovery #AIinBiotech #Biotech #GenerativeAI

Can Generative AI Design Antibodies Without a Lab?
A critical look at PG-AbD, a GFlowNet–PLM framework for antibody design, and why in silico metrics still need wet-lab validation
medium.com
April 16, 2025 at 7:00 PM
Can generative AI design antibodies without ever stepping into a lab?
I wrote about PG-AbD, a solid framework with no wet-lab validation, and why that's not a dead end, just a missed opportunity.
medium.com/@enginyapici...
#AI #DrugDiscovery #AntibodyDiscovery #AIinBiotech #Biotech #GenerativeAI
I wrote about PG-AbD, a solid framework with no wet-lab validation, and why that's not a dead end, just a missed opportunity.
medium.com/@enginyapici...
#AI #DrugDiscovery #AntibodyDiscovery #AIinBiotech #Biotech #GenerativeAI
What does “inactive” actually mean in drug discovery?
Most models are trained on actives, but real signal might lie in the compounds that quietly fail. I wrote about InertDB, a dataset of verified negatives, and what it means for model reliability.
tinyurl.com/InertDB-Medium
#DrugDiscovery #AI
Most models are trained on actives, but real signal might lie in the compounds that quietly fail. I wrote about InertDB, a dataset of verified negatives, and what it means for model reliability.
tinyurl.com/InertDB-Medium
#DrugDiscovery #AI

What Does “Inactive” Actually Mean in Drug Discovery?
A closer look at InertDB, a curated and AI-augmented resource for negative data
tinyurl.com
April 15, 2025 at 6:02 PM
What does “inactive” actually mean in drug discovery?
Most models are trained on actives, but real signal might lie in the compounds that quietly fail. I wrote about InertDB, a dataset of verified negatives, and what it means for model reliability.
tinyurl.com/InertDB-Medium
#DrugDiscovery #AI
Most models are trained on actives, but real signal might lie in the compounds that quietly fail. I wrote about InertDB, a dataset of verified negatives, and what it means for model reliability.
tinyurl.com/InertDB-Medium
#DrugDiscovery #AI
Reposted by Engin Yapici
CODA for the masses!
Valentina Matos-Romero, Ashley Kiemen and team have put together an ultra detailed protocol to use CODA for 3D single-cell mapping of tissues, organs, and organisms.
Use CODA by downloading this protocol here: www.biorxiv.org/content/10.1...
Valentina Matos-Romero, Ashley Kiemen and team have put together an ultra detailed protocol to use CODA for 3D single-cell mapping of tissues, organs, and organisms.
Use CODA by downloading this protocol here: www.biorxiv.org/content/10.1...
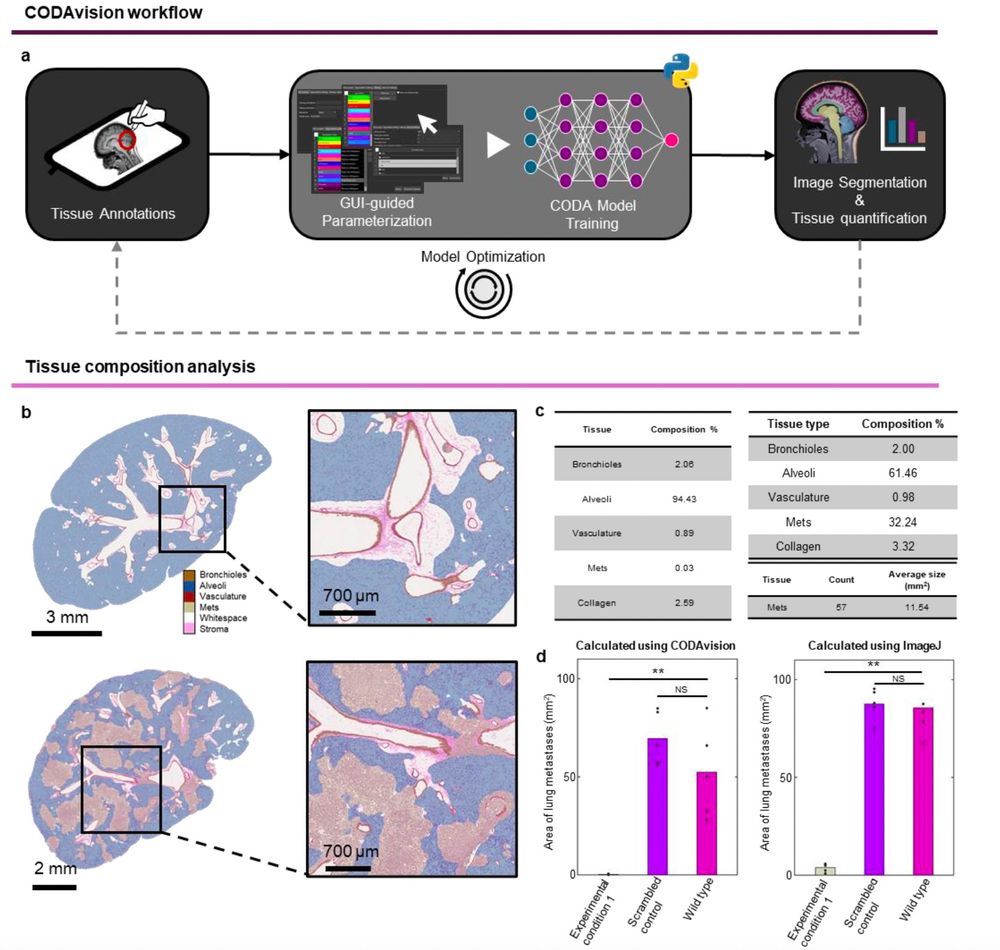
April 14, 2025 at 5:58 PM
CODA for the masses!
Valentina Matos-Romero, Ashley Kiemen and team have put together an ultra detailed protocol to use CODA for 3D single-cell mapping of tissues, organs, and organisms.
Use CODA by downloading this protocol here: www.biorxiv.org/content/10.1...
Valentina Matos-Romero, Ashley Kiemen and team have put together an ultra detailed protocol to use CODA for 3D single-cell mapping of tissues, organs, and organisms.
Use CODA by downloading this protocol here: www.biorxiv.org/content/10.1...
Reposted by Engin Yapici
Our @natrevimmunol.bsky.social review with @abhishekgarglab.bsky.social, @deadoc80.bsky.social and Kellie Smith is out!
We attempt to integrate the data on CD8 T cell dysfunction into a new framework of hypofunctionality in cancer and chronic infections!
www.nature.com/articles/s41...
We attempt to integrate the data on CD8 T cell dysfunction into a new framework of hypofunctionality in cancer and chronic infections!
www.nature.com/articles/s41...
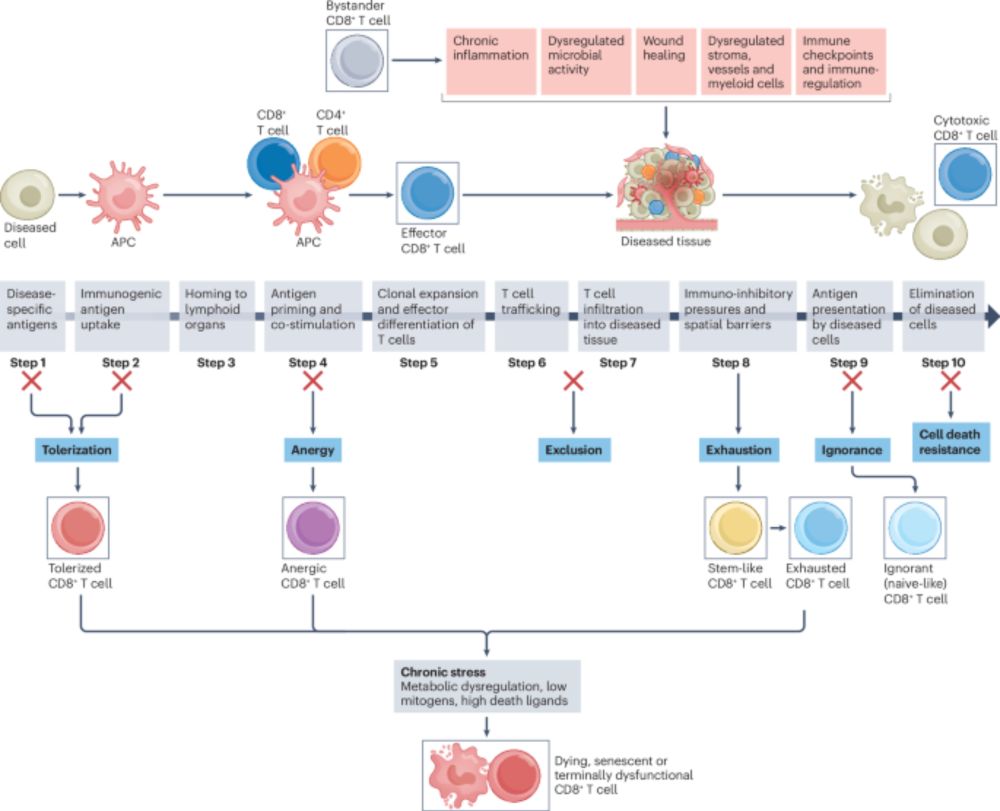
The diversity of CD8+ T cell dysfunction in cancer and viral infection - Nature Reviews Immunology
Beyond exhaustion, CD8+ T cells can adopt various dysfunctional states, including tolerant, anergic, senescent, ignorant and dying states, that compromise their ability to eradicate viruses or tumours...
www.nature.com
April 14, 2025 at 8:53 AM
Our @natrevimmunol.bsky.social review with @abhishekgarglab.bsky.social, @deadoc80.bsky.social and Kellie Smith is out!
We attempt to integrate the data on CD8 T cell dysfunction into a new framework of hypofunctionality in cancer and chronic infections!
www.nature.com/articles/s41...
We attempt to integrate the data on CD8 T cell dysfunction into a new framework of hypofunctionality in cancer and chronic infections!
www.nature.com/articles/s41...
First post covers a microwell platform (MoSMAR-chip) that screens for antigen specificity, function, and transcriptomics, single-cell, high-throughput, no droplet systems.
medium.com/@enginyapici...
#SingleCell #AntibodyDiscovery #ScreeningTech #FunctionalAssays #Biotech
medium.com/@enginyapici...
#SingleCell #AntibodyDiscovery #ScreeningTech #FunctionalAssays #Biotech

A High-Throughput Platform for Single-Cell Antibody Discovery: Inside the MoSMAR-Chip
A microwell-based approach to link antibody function, specificity, and transcriptional state in LLPCs and MBCs
medium.com
April 13, 2025 at 9:23 PM
First post covers a microwell platform (MoSMAR-chip) that screens for antigen specificity, function, and transcriptomics, single-cell, high-throughput, no droplet systems.
medium.com/@enginyapici...
#SingleCell #AntibodyDiscovery #ScreeningTech #FunctionalAssays #Biotech
medium.com/@enginyapici...
#SingleCell #AntibodyDiscovery #ScreeningTech #FunctionalAssays #Biotech
I started a Medium series on tools and technologies in drug discovery, especially antibody development, screening, and MoA assays. I’ll be breaking down papers that offer something useful (or not).
Intro: medium.com/@enginyapici...
#DrugDiscovery #AntibodyEngineering #Bioassays #MoA #Biotech
Intro: medium.com/@enginyapici...
#DrugDiscovery #AntibodyEngineering #Bioassays #MoA #Biotech

From Assay to Algorithm: A Scientist’s Perspective on What’s Worth Watching
A scientist’s guide to high-throughput screening, phenotypic assays, antibody discovery, and cutting-edge drug development tools
medium.com
April 13, 2025 at 9:23 PM
I started a Medium series on tools and technologies in drug discovery, especially antibody development, screening, and MoA assays. I’ll be breaking down papers that offer something useful (or not).
Intro: medium.com/@enginyapici...
#DrugDiscovery #AntibodyEngineering #Bioassays #MoA #Biotech
Intro: medium.com/@enginyapici...
#DrugDiscovery #AntibodyEngineering #Bioassays #MoA #Biotech
Hi, I’m Engin. I work in drug discovery, mostly antibody discovery and development, high-throughput screening, and MoA functional assays. I’m here to share what I’ve learned, what I’m still figuring out, and to learn from others thinking deeply about how we move this field forward.
April 13, 2025 at 9:21 PM
Hi, I’m Engin. I work in drug discovery, mostly antibody discovery and development, high-throughput screening, and MoA functional assays. I’m here to share what I’ve learned, what I’m still figuring out, and to learn from others thinking deeply about how we move this field forward.

