
It is the first deep-learning-based ("AI") drop-in replacement for TrackMate's regular LAP-Track algorithm.
Tracks out of the box for many types of datasets (bacteria, fluorescent nuclei, phase contrast cell culture etc), zero parameters to tune 🤖
It is the first deep-learning-based ("AI") drop-in replacement for TrackMate's regular LAP-Track algorithm.
Tracks out of the box for many types of datasets (bacteria, fluorescent nuclei, phase contrast cell culture etc), zero parameters to tune 🤖
HUGE shoutout to Aditi from @bioimagingna.bsky.social for being the organizational wizard!
See you in the recordings and at next year's!
HUGE shoutout to Aditi from @bioimagingna.bsky.social for being the organizational wizard!
See you in the recordings and at next year's!
www.cell.com/cell/fulltex...

www.cell.com/cell/fulltex...



wi.mit.edu/news/alterna...

wi.mit.edu/news/alterna...
(source: doi.org/10.48550/arX...)

(source: doi.org/10.48550/arX...)
Wishing you'd submitted a tool or workshop but missed the deadline? GREAT NEWS, we just extended one week! Submit by Friday - airtable.com/app2zpB8d82r...
Got an idea for an image analysis workshop? Share it at Halfway to I2K: Online Tutorials on Image Analysis (Nov 17–19, 2025), held with Zoom Events!
Submit by Oct 31: buff.ly/lMhK4vz
Website: buff.ly/QP36RZB
Register for the workshops: buff.ly/8nNAI18

Wishing you'd submitted a tool or workshop but missed the deadline? GREAT NEWS, we just extended one week! Submit by Friday - airtable.com/app2zpB8d82r...
Got an idea for an image analysis workshop? Share it at Halfway to I2K: Online Tutorials on Image Analysis (Nov 17–19, 2025), held with Zoom Events!
Submit by Oct 31: buff.ly/lMhK4vz
Website: buff.ly/QP36RZB
Register for the workshops: buff.ly/8nNAI18

🎓 Courses, Seminars, Round Tables, Workshops. The overall theme concerns #biological_imaging in its most interdisciplinary aspects.
👉 Program: imabio-cnrs.fr
#GDRImaBio #Mifobio2025

🎓 Courses, Seminars, Round Tables, Workshops. The overall theme concerns #biological_imaging in its most interdisciplinary aspects.
👉 Program: imabio-cnrs.fr
#GDRImaBio #Mifobio2025
My workflow will be to autotrace and then manually correct the autotraces, reducing the time for our tracing significantly!
My workflow will be to autotrace and then manually correct the autotraces, reducing the time for our tracing significantly!
If you make image analysis software and want to teach it, workshop submissions are open now! We'd love to have your tool highlighted.
#microscopycommunity- want to learn open source image analysis or share your knowledge to help others? We’ve got a FREE virtual workshop Nov 17-19! Now accepting workshop session applications!
Learn more & sign up: buff.ly/esGIotD
If you make image analysis software and want to teach it, workshop submissions are open now! We'd love to have your tool highlighted.
Kuffer & Marzilli engineered conditionally stable MS2 & PP7 coat proteins (dMCP & dPCP) that degrade unless bound to RNA, enabling ultra–low-background, single-mRNA imaging in live cells.
🔗 www.nature.com/articles/s41...
🧬 www.addgene.org/John_Ngo/
Kuffer & Marzilli engineered conditionally stable MS2 & PP7 coat proteins (dMCP & dPCP) that degrade unless bound to RNA, enabling ultra–low-background, single-mRNA imaging in live cells.
🔗 www.nature.com/articles/s41...
🧬 www.addgene.org/John_Ngo/

Finished my PhD on glucose signaling in yeast's G2 phase.
Thanks to the whole Ewald lab and our fantastic collaborators!


Finished my PhD on glucose signaling in yeast's G2 phase.
Thanks to the whole Ewald lab and our fantastic collaborators!
doi.org/10.1101/2025...
Like ❤️, repost 🔂, and most importantly... please send feedback ✉️ our way! 🙏

doi.org/10.1101/2025...
Like ❤️, repost 🔂, and most importantly... please send feedback ✉️ our way! 🙏
www.nature.com/articles/s41...
rdcu.be/epIB7
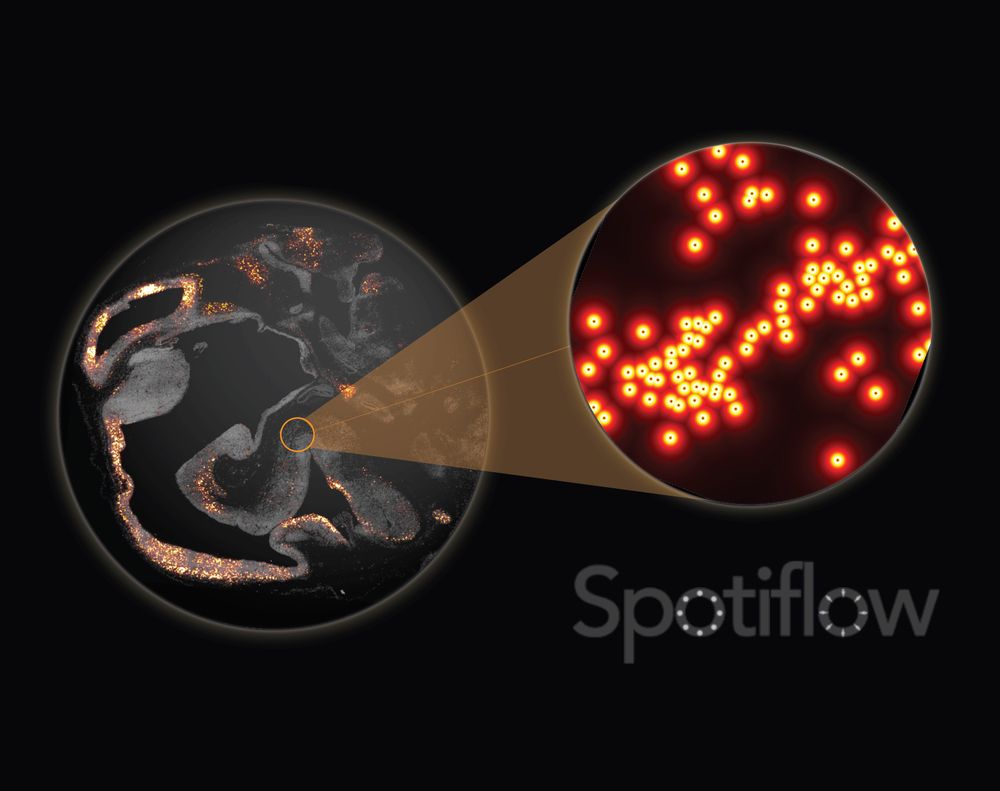
www.nature.com/articles/s41...
rdcu.be/epIB7
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)
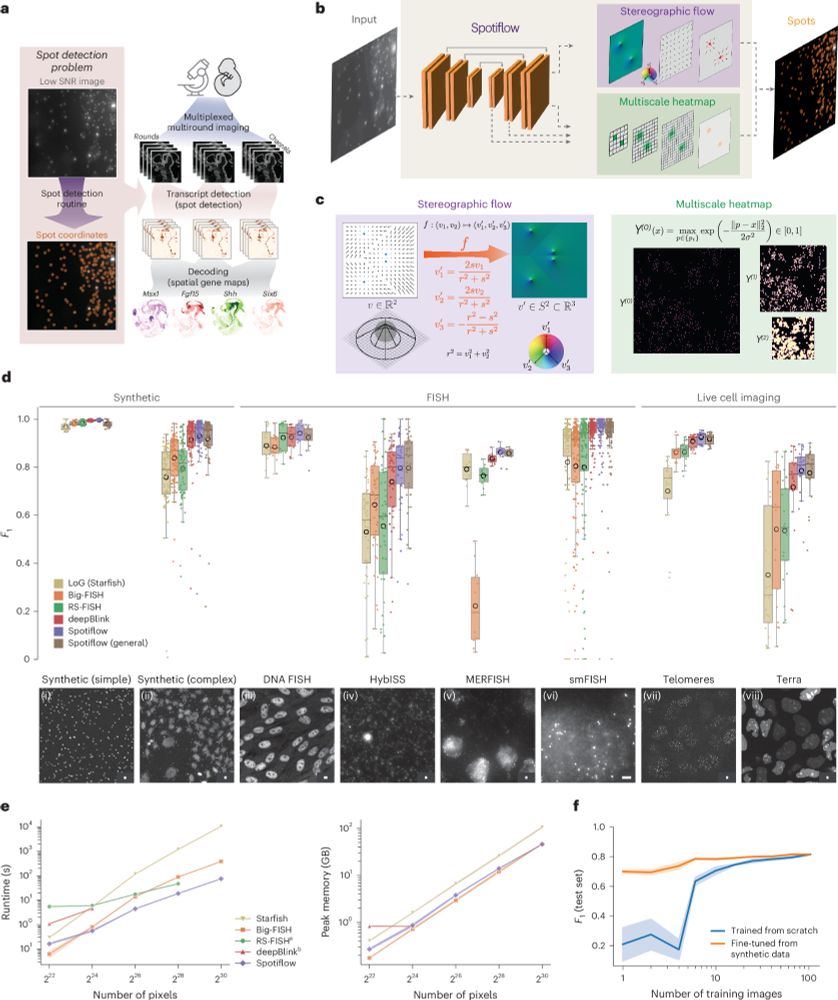
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)
nimbusimage.com

nimbusimage.com
Day 1️⃣: Workshops for life scientists, software devs, AI researchers & image analysts
Day 2️⃣: Talks, discussions & networking
If that sounds like you, don’t miss it!
👉 ai4life.eurobioimaging.eu/event/ai4lif...
Day 1️⃣: Workshops for life scientists, software devs, AI researchers & image analysts
Day 2️⃣: Talks, discussions & networking
If that sounds like you, don’t miss it!
👉 ai4life.eurobioimaging.eu/event/ai4lif...
github.com/haesleinhuep...
github.com/haesleinhuep...


