PhD in Bioinformatics from Temple University
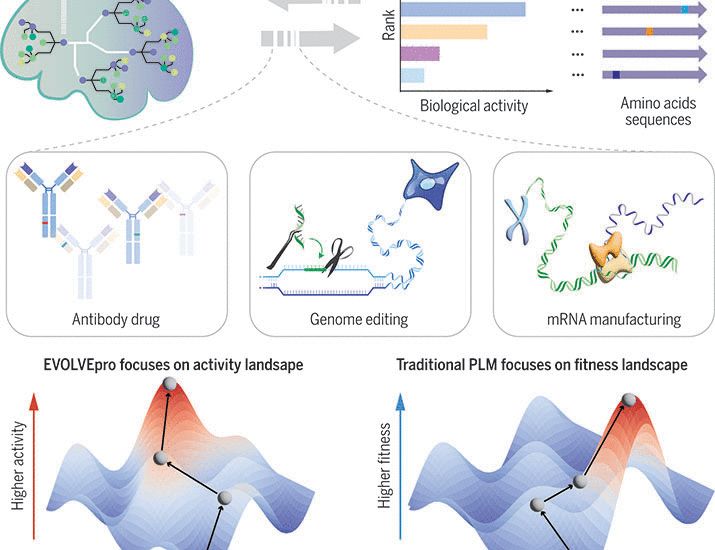
[1/N]
www.biorxiv.org/content/10.1...

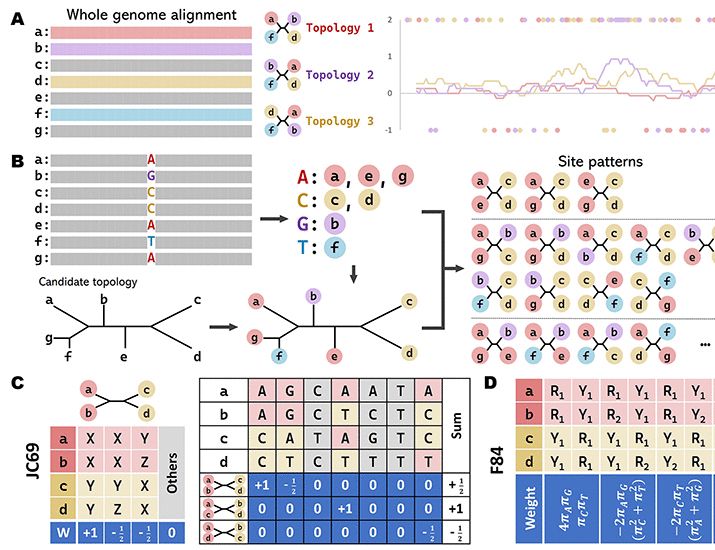
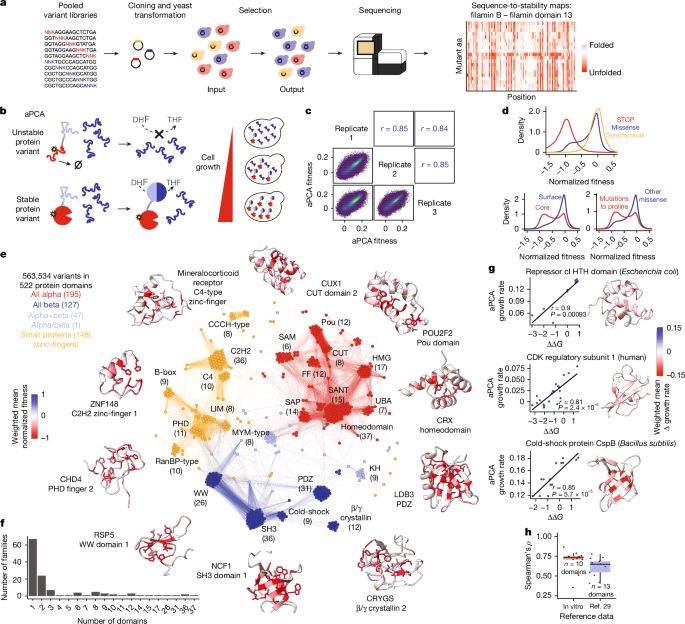
[1/N]
www.biorxiv.org/content/10.1...
More info here: molevolworkshop.github.io
Apply here: www.mbl.edu/education/ad...
Application deadline: Jan. 29th
Workshop dates: May 22nd - June 1st

More info here: molevolworkshop.github.io
Apply here: www.mbl.edu/education/ad...
Application deadline: Jan. 29th
Workshop dates: May 22nd - June 1st