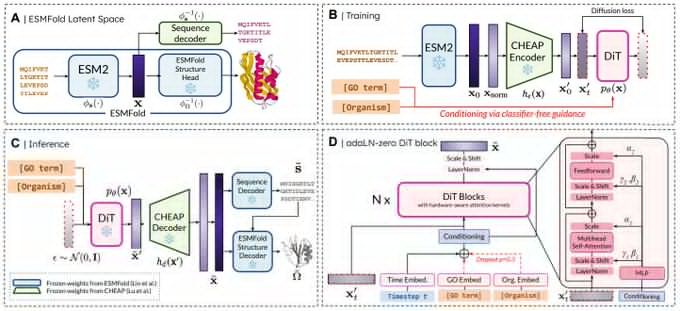
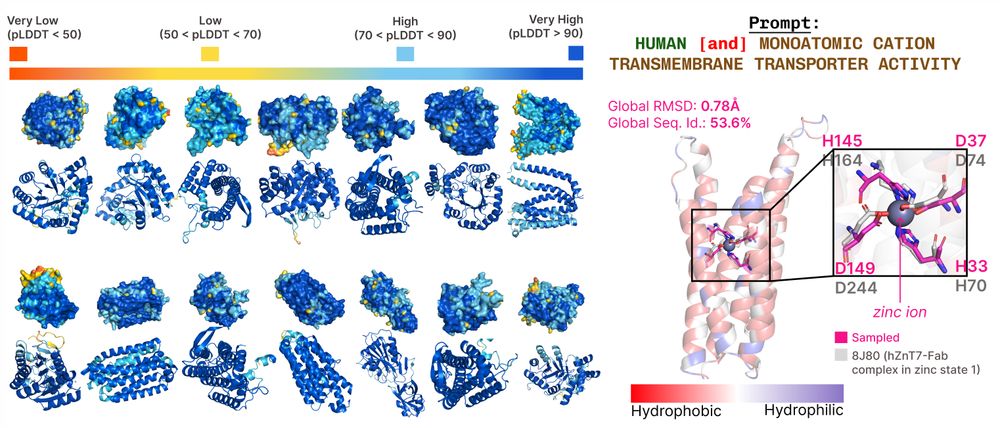
Amy Lu
@amyxlu.bsky.social
870 followers
200 following
10 posts
CS PhD Student at UC Berkeley & AI for drug discovery at Prescient Design 🇨🇦
Posts
Media
Videos
Starter Packs
Reposted by Amy Lu
Reposted by Amy Lu
Amy Lu
@amyxlu.bsky.social
· Dec 15
Amy Lu
@amyxlu.bsky.social
· Dec 10
Reposted by Amy Lu
Amy Lu
@amyxlu.bsky.social
· Dec 6