Ana Fiszbein
@anafiszbein.bsky.social
82 followers
25 following
18 posts
Molecular and computational biologist. Assistant professor at Boston University in Biology and Computing & Data Sciences. Argentinian
www.fiszbeinlab.com
Posts
Media
Videos
Starter Packs
Ana Fiszbein
@anafiszbein.bsky.social
· May 16
Ana Fiszbein
@anafiszbein.bsky.social
· May 16

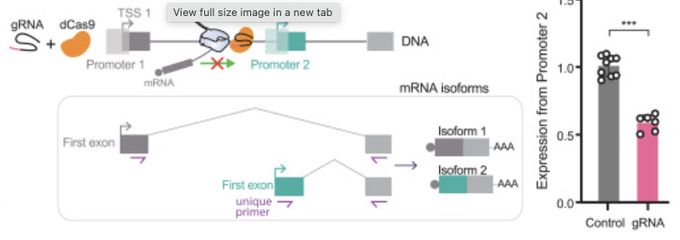
U1 snRNP regulates alternative promoter activity by inhibiting premature polyadenylation
Kim et al. uncover a role for U1 snRNP in regulating internal promoter activity. Beyond
its canonical role in splicing, U1 snRNP suppresses premature polyadenylation, enabling
upstream transcription t...
www.cell.com
Ana Fiszbein
@anafiszbein.bsky.social
· Jan 16
Ana Fiszbein
@anafiszbein.bsky.social
· Jan 16

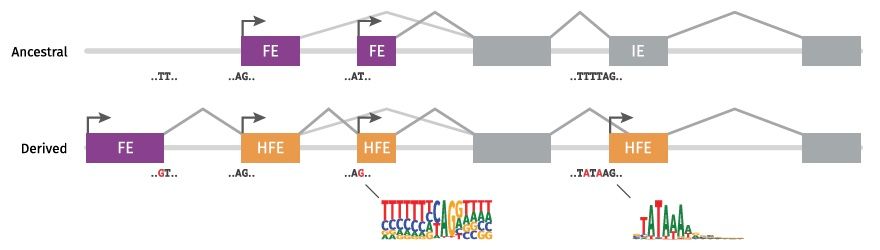
Hybrid exons evolved by coupling transcription initiation and splicing at the nucleotide level
Abstract. Exons within transcripts are traditionally classified as first, internal or last exons, each governed by different regulatory mechanisms. We rece
academic.oup.com
Ana Fiszbein
@anafiszbein.bsky.social
· Jan 16