transformer)‑LSTM. Details: www.nature.com/articles/s41...
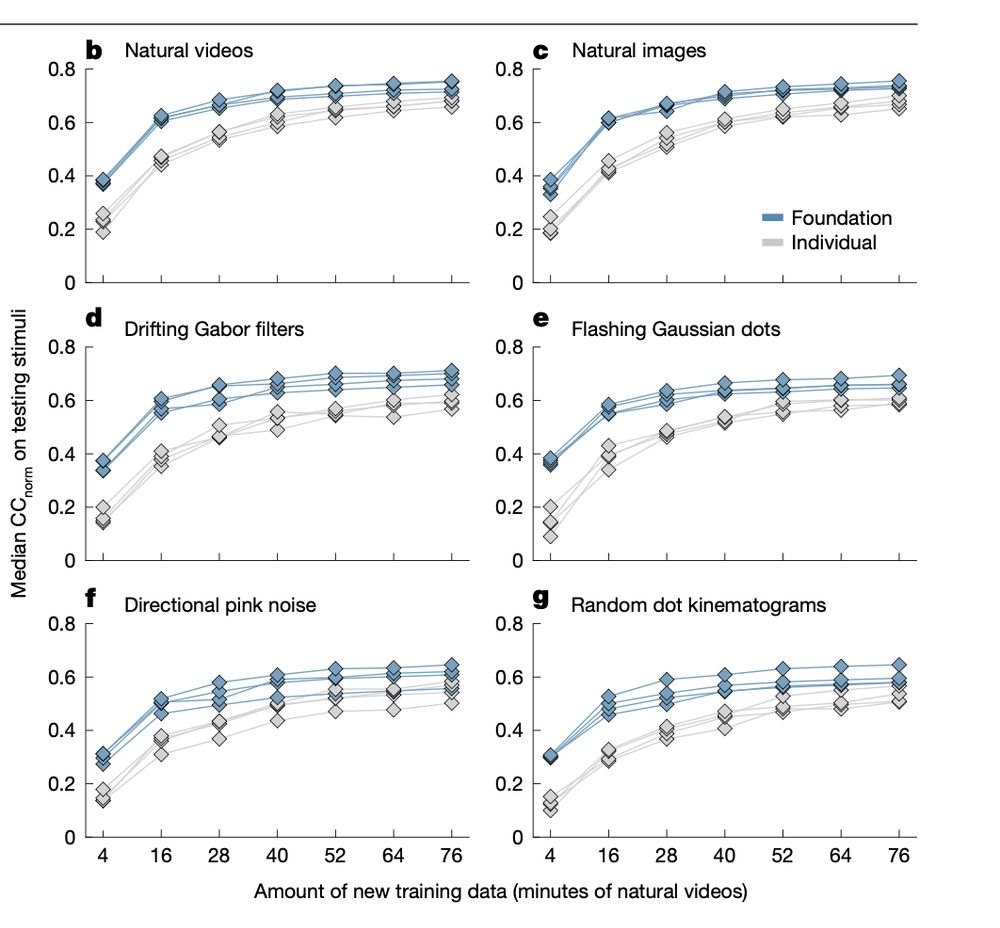
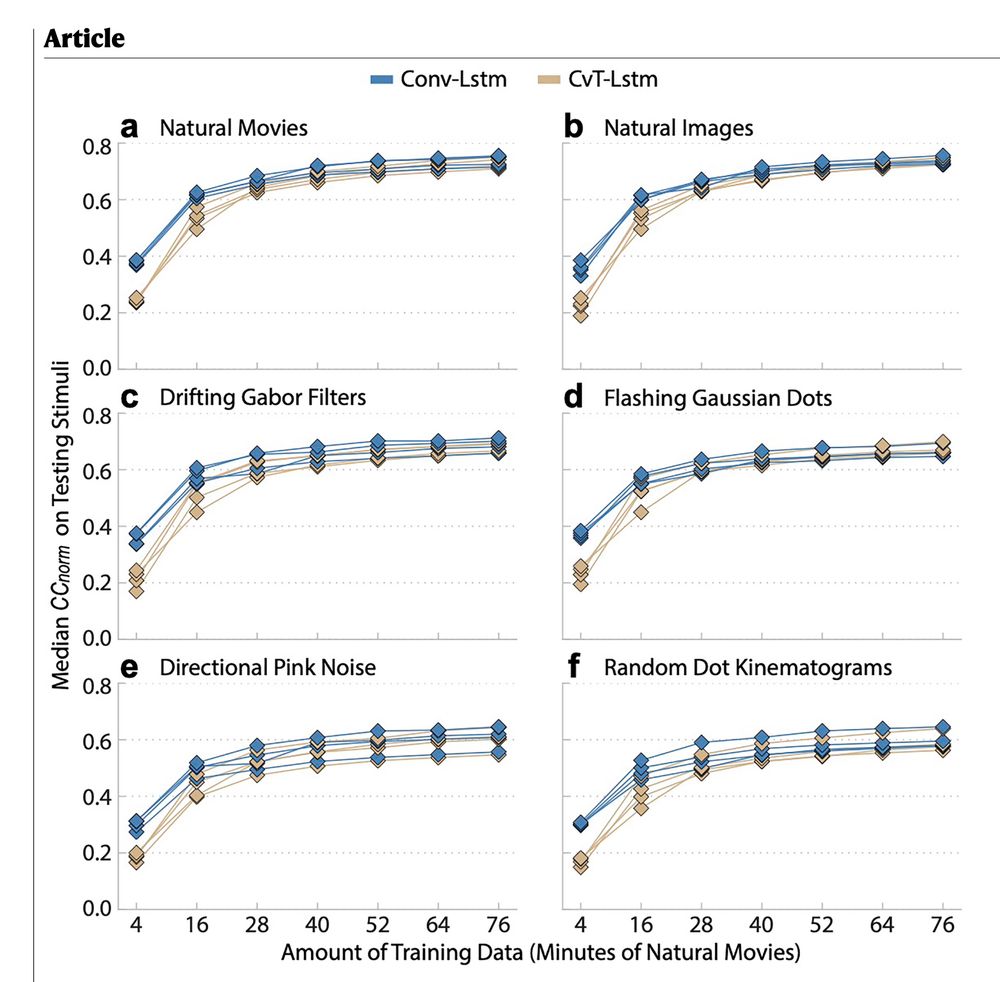
transformer)‑LSTM. Details: www.nature.com/articles/s41...


Apply here: www.linkedin.com/jobs/view/42...
Or email us at [email protected]

Apply here: www.linkedin.com/jobs/view/42...
Or email us at [email protected]
www.biorxiv.org/content/bior...
www.biorxiv.org/content/bior...
www.biorxiv.org/content/bior...
www.biorxiv.org/content/bior...
@stanforduniversity.bsky.social @stanfordmedicine.bsky.social @BCM @Allen @Princeton @unigoettingen.bsky.social
#MICrONS #NeuroAI #Connectomics #FoundationModels #AI
@stanforduniversity.bsky.social @stanfordmedicine.bsky.social @BCM @Allen @Princeton @unigoettingen.bsky.social
#MICrONS #NeuroAI #Connectomics #FoundationModels #AI

