Spanish National Cancer Research Centre (CNIO)
Send us an email via the adresses on the website and we will investigate.
Send us an email via the adresses on the website and we will investigate.
And that winds up our work in @gencodegenes.bsky.social
It has been fun.
Work carried out by Miguel Maquedano and @danielcerdan.bsky.social

And that winds up our work in @gencodegenes.bsky.social
It has been fun.
Work carried out by Miguel Maquedano and @danielcerdan.bsky.social
We merged and compared @ensembl.org / @gencodegenes.bsky.social , RefSeq and UniProtKB coding genes and investigated the agreements and discrepancies.
Details of what we found in the thread ...

We merged and compared @ensembl.org / @gencodegenes.bsky.social , RefSeq and UniProtKB coding genes and investigated the agreements and discrepancies.
Details of what we found in the thread ...
Because of recent changes to government funding, the NIH grant that supported FlyBase has been "terminated." flybase.org

Because of recent changes to government funding, the NIH grant that supported FlyBase has been "terminated." flybase.org
Principal isoforms for a total of 73 human genes have been updated manually.
Principal isoforms for a total of 73 human genes have been updated manually.
📍Organizan: @saludisciii.bsky.social @cniostopcancer.bsky.social @bsc-cns.bsky.social
🤝 Colaboran: SEBiot,Instituto Roche
📅 Plazas limitadas
🔗 masterbioinformatica.com

📍Organizan: @saludisciii.bsky.social @cniostopcancer.bsky.social @bsc-cns.bsky.social
🤝 Colaboran: SEBiot,Instituto Roche
📅 Plazas limitadas
🔗 masterbioinformatica.com
These principal isoforms are exclusive to the human gene set and are principal isoforms that have been manually corrected. In GENCODE v47 there were 45 genes with Principal:M isoforms.
Here is DCD:

These principal isoforms are exclusive to the human gene set and are principal isoforms that have been manually corrected. In GENCODE v47 there were 45 genes with Principal:M isoforms.
Here is DCD:


In particular it shows that what was previously thought to be the human WASH1C gene is actually a pseudogene.
bsky.app/profile/mich...
This goes some way to explaining why so little is known about this vital gene, an integral part of the WASH complex.
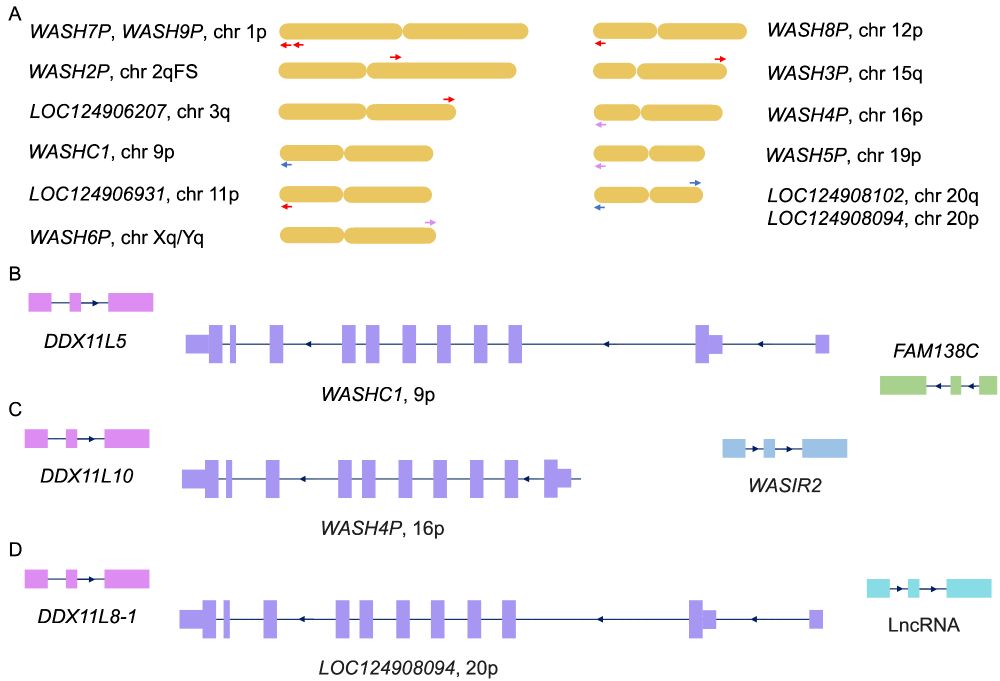
In particular it shows that what was previously thought to be the human WASH1C gene is actually a pseudogene.
bsky.app/profile/mich...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

¡Para celebrar que hoy es el Doctor Who Day
sorteamos una colección completa de los cómics que hemos publicado del Doctor, es decir, 11 TEBEOS!
👉 Síguenos
👉 Comparte este maripost. Retuitea este skeet. Ya sabes lo que quiero
👉 ¡El sorteo termina a las 23:59 del 25/11!
¡¡SUERTE!!

¡Para celebrar que hoy es el Doctor Who Day
sorteamos una colección completa de los cómics que hemos publicado del Doctor, es decir, 11 TEBEOS!
👉 Síguenos
👉 Comparte este maripost. Retuitea este skeet. Ya sabes lo que quiero
👉 ¡El sorteo termina a las 23:59 del 25/11!
¡¡SUERTE!!
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

Agreement with MANE Select transcripts in human (no MANE Select in mouse) has risen slightly to 17,661 genes.
appris.bioinfo.cnio.es#/
Agreement with MANE Select transcripts in human (no MANE Select in mouse) has risen slightly to 17,661 genes.
appris.bioinfo.cnio.es#/
The key takeaways are that it is common and that most translation from 5' UTR is in-frame, so the effect at the protein level is to extend the N-terminal.
academic.oup.com/nar/advance-...

The key takeaways are that it is common and that most translation from 5' UTR is in-frame, so the effect at the protein level is to extend the N-terminal.
academic.oup.com/nar/advance-...

