Jun Ishigohoka
@junishigohoka.bsky.social
180 followers
220 following
33 posts
Evolutionary biologist
Tübingen < Plön < Kiel < Sapporo < Tokyo
https://junishigohoka.github.io/
Posts
Media
Videos
Starter Packs
Pinned
Jun Ishigohoka
@junishigohoka.bsky.social
· Jan 27
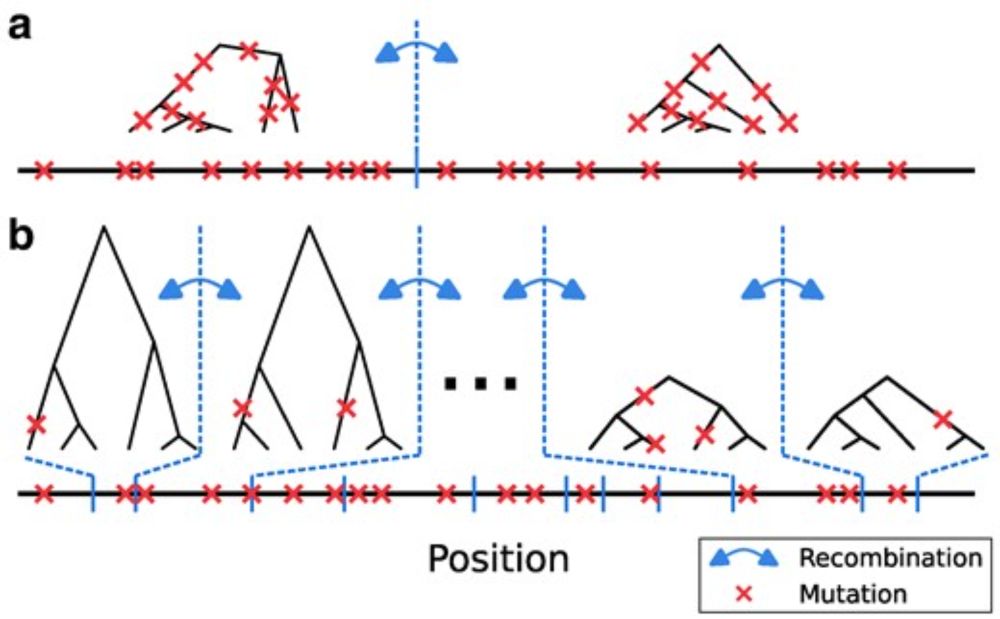
High-recombining genomic regions affect demography inference based on ancestral recombination graphs
Abstract. Multiple methods of demography inference are based on the ancestral recombination graph. This powerful approach uses observed mutations to model
doi.org
Reposted by Jun Ishigohoka
Reposted by Jun Ishigohoka
Reposted by Jun Ishigohoka
Reposted by Jun Ishigohoka
Reposted by Jun Ishigohoka
Reposted by Jun Ishigohoka
Reposted by Jun Ishigohoka
Reposted by Jun Ishigohoka
Reposted by Jun Ishigohoka
Reposted by Jun Ishigohoka
James Kitchens
@kitchensjn.bsky.social
· Aug 19

tskit_arg_visualizer: interactive plotting of ancestral recombination graphs
Summary: Ancestral recombination graphs (ARGs) are a complete representation of the genetic relationships between recombining lineages and are of central importance in population genetics. Recent brea...
arxiv.org
Reposted by Jun Ishigohoka
Reposted by Jun Ishigohoka
Jun Ishigohoka
@junishigohoka.bsky.social
· Aug 16

dArc1 controls sugar reward valuation in Drosophila melanogaster
Bervoets et al. show that the retrotransposon-derived gene dArc1 is expressed in serotonergic
neurons of the Drosophila brain. They found that loss of dArc leads to sugar reward
overvaluation during l...
www.cell.com
Jun Ishigohoka
@junishigohoka.bsky.social
· Aug 14

The genomic origin of the unique chaetognath body plan - Nature
Genomic, single-cell transcriptomic and epigenetic analyses show that chaetognaths, following extensive gene loss in the gnathiferan lineage, relied on newly evolved genes and lineage-specific tandem ...
www.nature.com
Reposted by Jun Ishigohoka