https://mkanai.github.io/

e.g., TNRC18 intron variant (114x enriched) is a risk factor for IBD but protective against autoimmune hypothyroidism.

e.g., TNRC18 intron variant (114x enriched) is a risk factor for IBD but protective against autoimmune hypothyroidism.


Full cascade variants (caQTL + eQTL + Link) show 2x higher GWAS colocalization rates compared to those affecting chromatin alone, establishing a clear hierarchy for prioritizing causal variants.

Full cascade variants (caQTL + eQTL + Link) show 2x higher GWAS colocalization rates compared to those affecting chromatin alone, establishing a clear hierarchy for prioritizing causal variants.




We profiled ~10M PBMCs (snRNA-seq + snATAC-seq) from 1,108 Finnish donors to map how genetic variants drive complex disease through chromatin and gene regulation 🧵👇
🔗 Link: www.medrxiv.org/content/10.1...
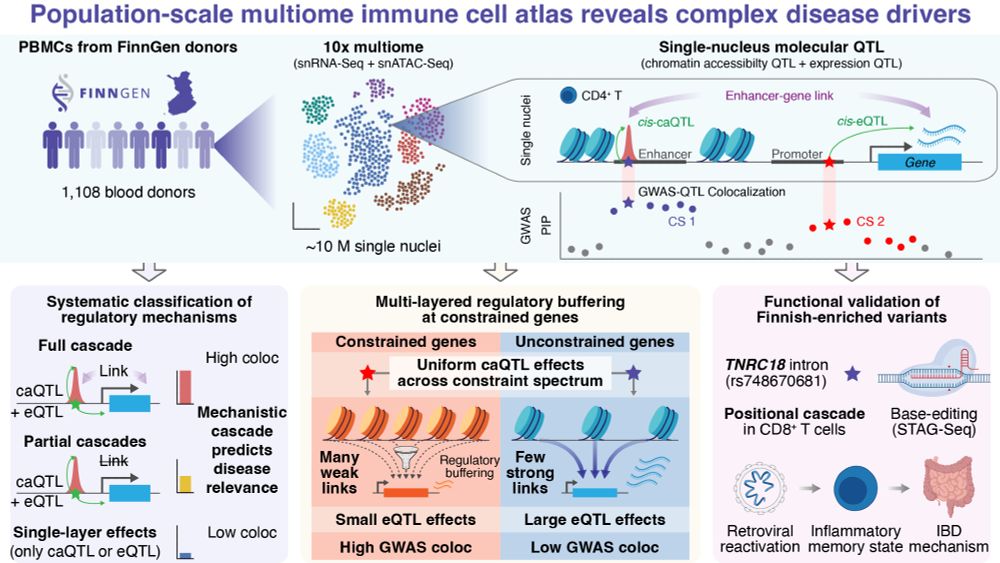
We profiled ~10M PBMCs (snRNA-seq + snATAC-seq) from 1,108 Finnish donors to map how genetic variants drive complex disease through chromatin and gene regulation 🧵👇
🔗 Link: www.medrxiv.org/content/10.1...

