Micha Müller
@michamuller.bsky.social
290 followers
890 following
17 posts
PhD student in the Tanenbaum lab at the Hubrecht Institute. Previously in the Pelkmans lab at UZH.
Interested in quantitative (live-cell) imaging, single-cell barcoding technologies, single-cell Omics, virus-host competition and many other things.
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Micha Müller
Jop Kind
@jopkind.bsky.social
· Jul 10
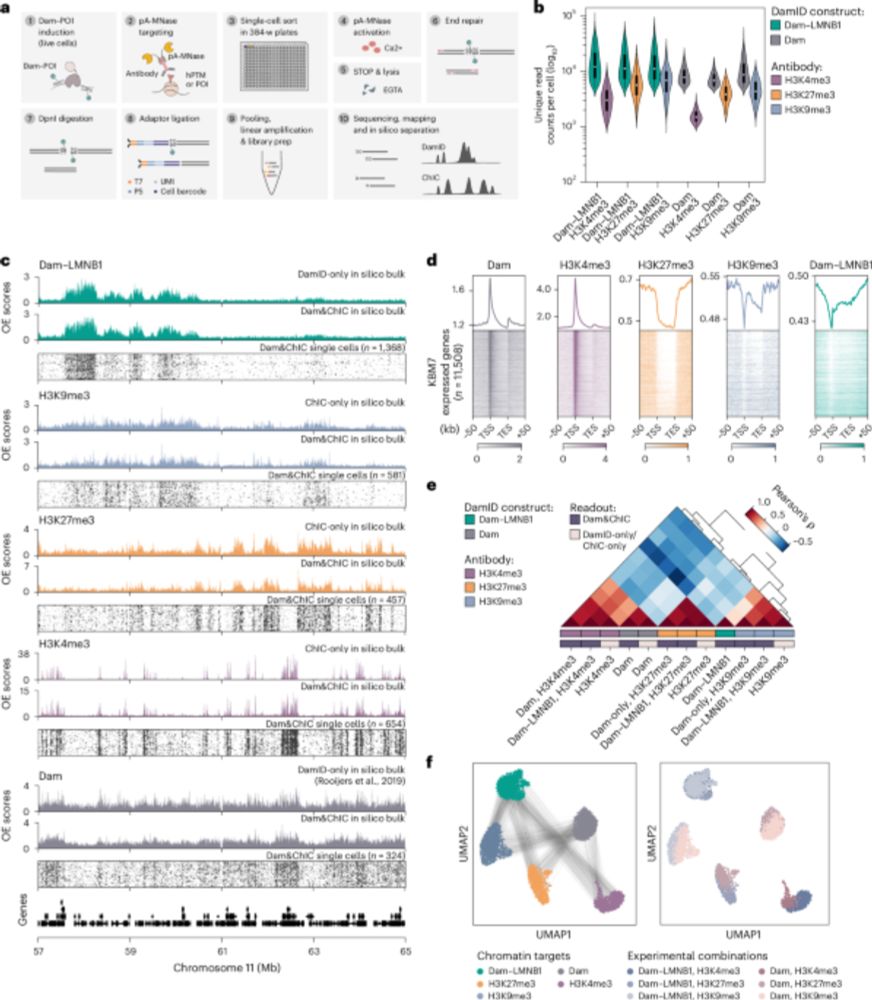
Retrospective and multifactorial single-cell profiling reveals sequential chromatin reorganization during X inactivation - Nature Cell Biology
Kefalopoulou, Rullens et al. develop Dam&ChIC to assay chromatin state at two different time points in the same cell. The method was used to study the reorganization of LADs during cell division a...
www.nature.com
Reposted by Micha Müller
Reposted by Micha Müller
Reposted by Micha Müller
Micha Müller
@michamuller.bsky.social
· Jan 21
Micha Müller
@michamuller.bsky.social
· Jan 21
Micha Müller
@michamuller.bsky.social
· Jan 21
Micha Müller
@michamuller.bsky.social
· Jan 21
Micha Müller
@michamuller.bsky.social
· Jan 21
Micha Müller
@michamuller.bsky.social
· Jan 21
Micha Müller
@michamuller.bsky.social
· Jan 21
Micha Müller
@michamuller.bsky.social
· Jan 21

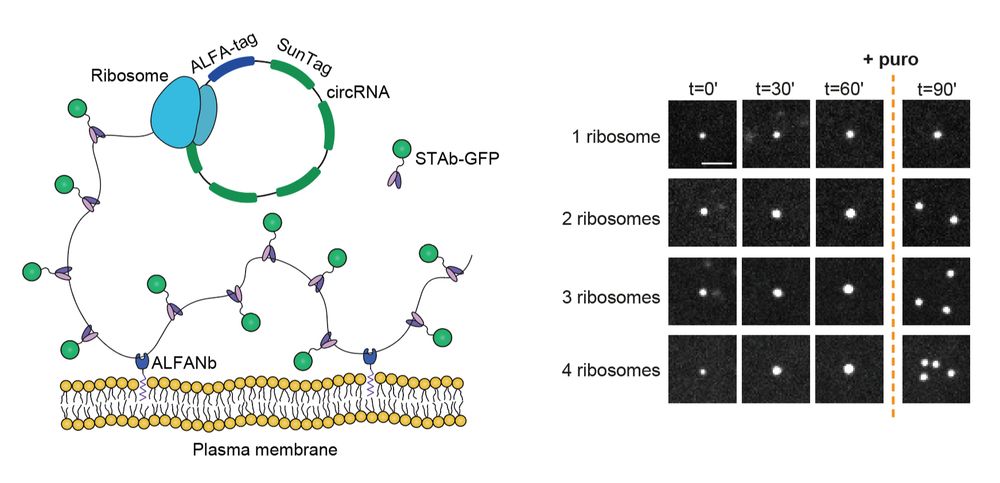
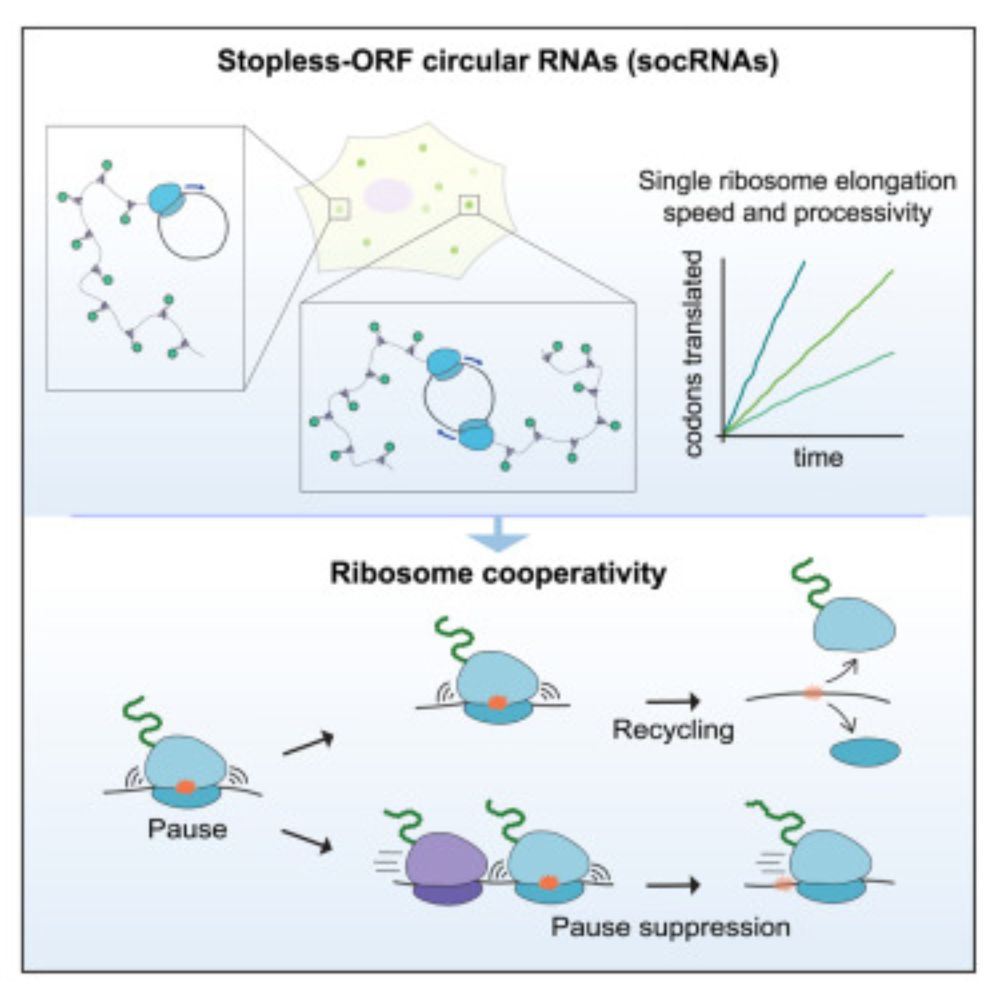
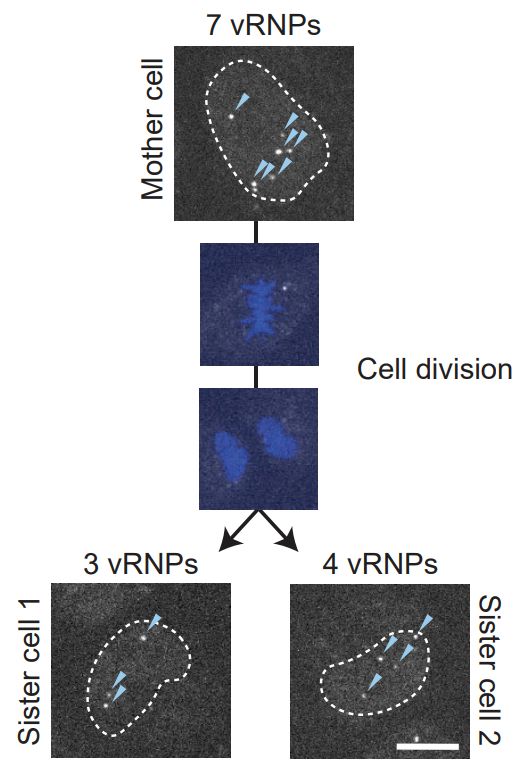
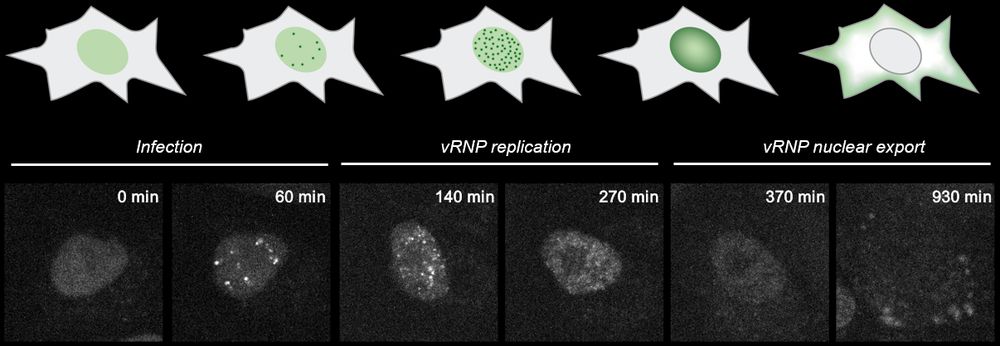
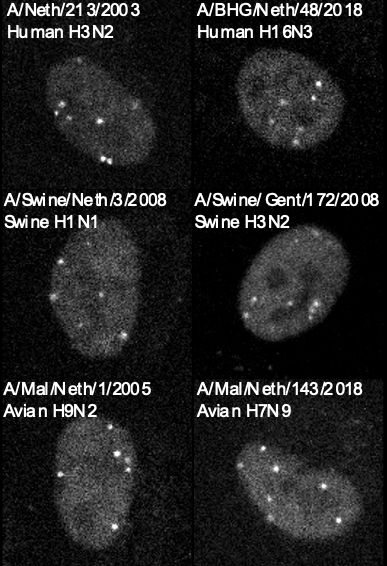
Mapping the complete influenza A virus infection cycle through single vRNP imaging
Cell-to-cell heterogeneity is a common feature of viral infection that can generate enormous complexity, complicating understanding of infection progression and interpretation of differences between v...
www.biorxiv.org
Reposted by Micha Müller
Micha Müller
@michamuller.bsky.social
· Nov 20