@ruhrsynapse.bsky.social
27 followers
63 following
30 posts
Neuroscientist, human genetics @UK Essen
Views are their own.
Posts
Media
Videos
Starter Packs
Pinned
Reposted
Yang Luo
@y-luo.bsky.social
· May 25
Reposted
Reposted
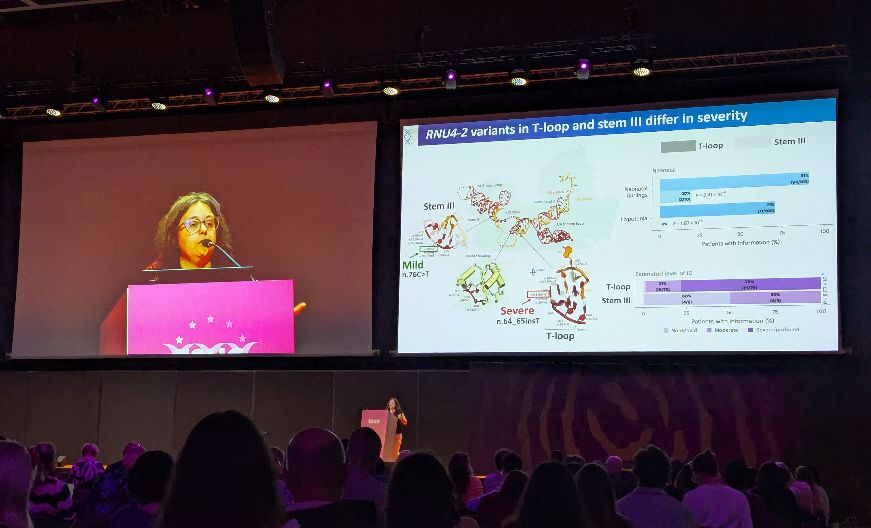
Nicky Whiffin
@nickywhiffin.bsky.social
· May 26
Reposted
Reposted
Reposted
Reposted
Reposted
Reposted
Nicky Whiffin
@nickywhiffin.bsky.social
· May 25
Reposted
Reposted
Reposted
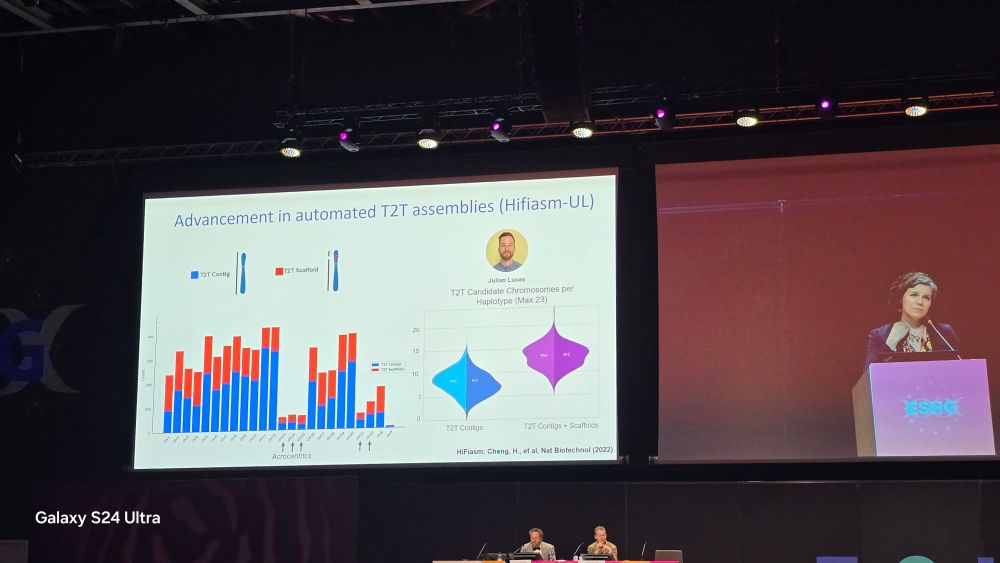
Alex Hoischen
@ahoischen.bsky.social
· May 24
Reposted
Reposted
Reposted