Sameed Siddiqui
@sameedms.bsky.social
41 followers
35 following
17 posts
Californian lost in the Northeast ☀️.
PhD @ MIT Computational and Systems Biology | MBA Fellow at MIT Sloan. @SabetiLab member
Posts
Media
Videos
Starter Packs
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21

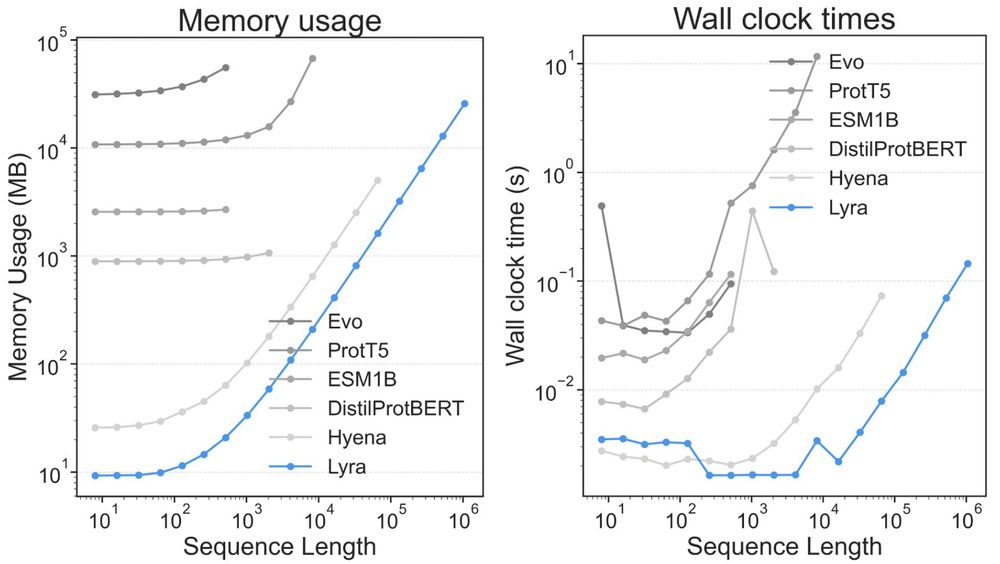
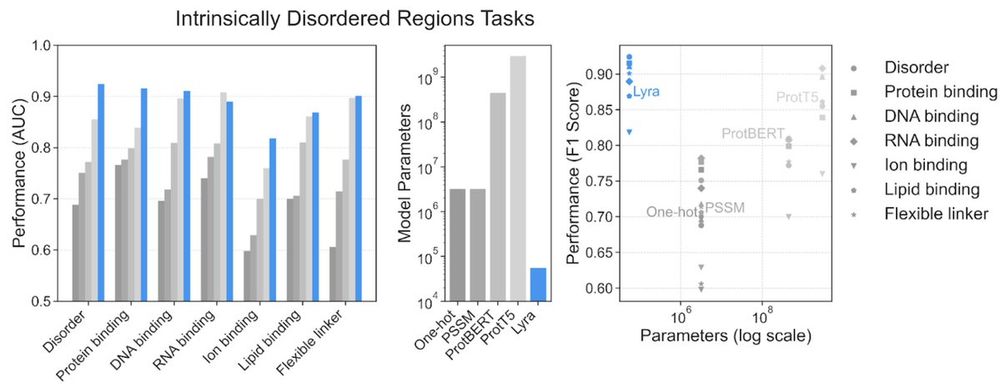
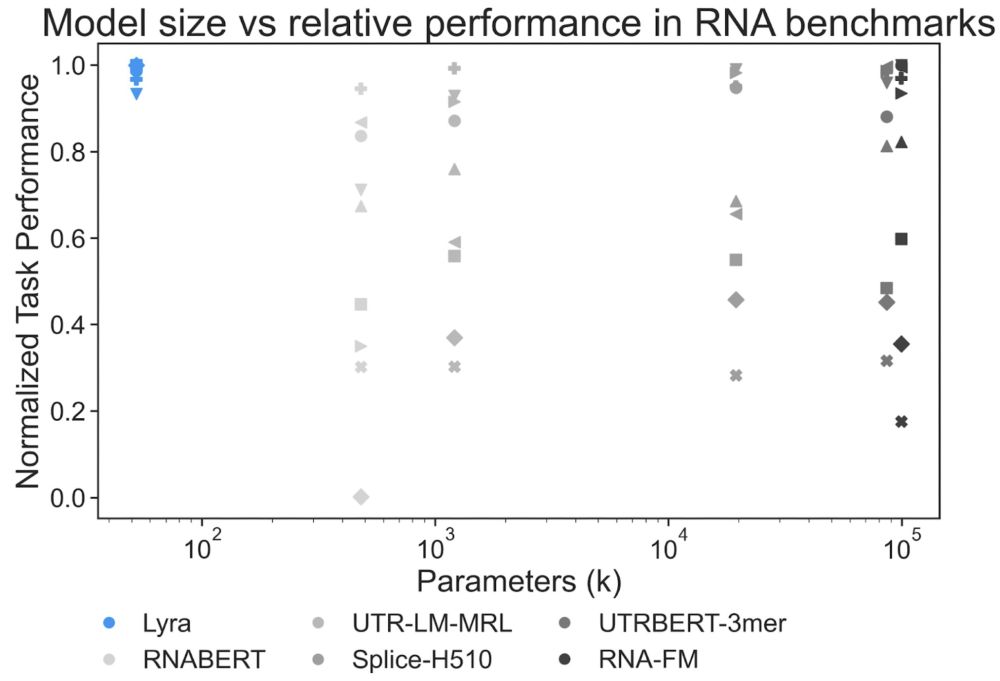
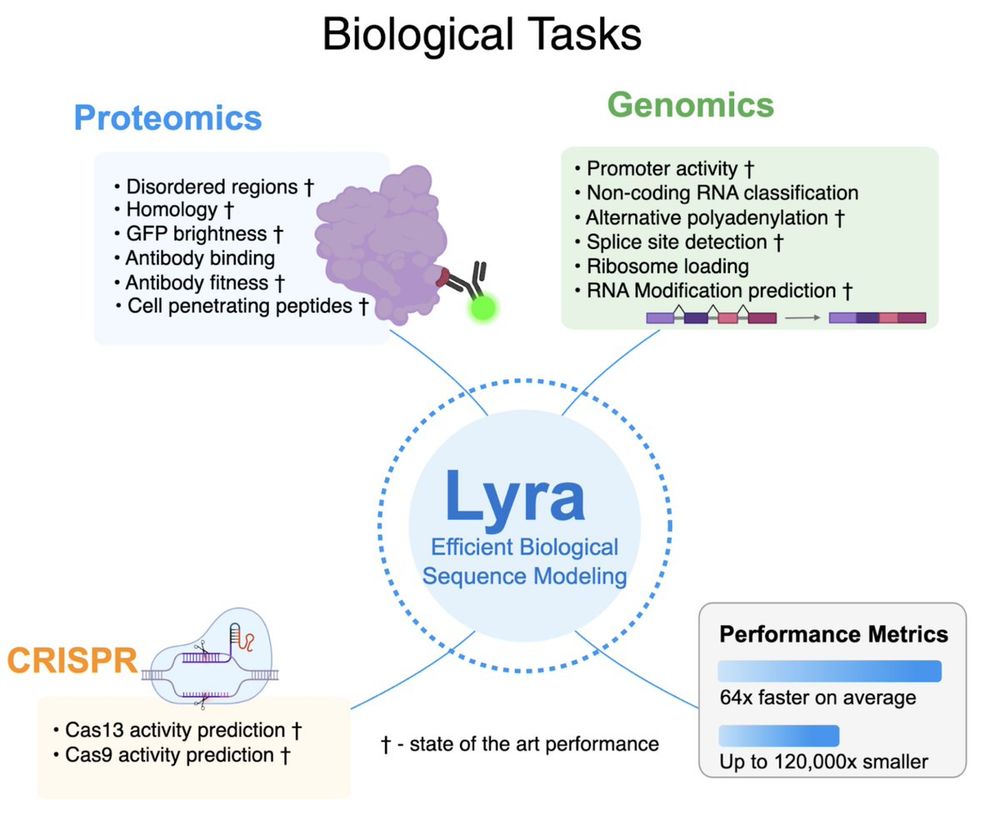
Lyra: An Efficient and Expressive Subquadratic Architecture for Modeling Biological Sequences
Deep learning architectures such as convolutional neural networks and Transformers have revolutionized biological sequence modeling, with recent advances driven by scaling up foundation and task-speci...
arxiv.org
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21
Sameed Siddiqui
@sameedms.bsky.social
· Mar 21