Stefano Mangiola
@stemang.bsky.social
1.8K followers
24 following
41 posts
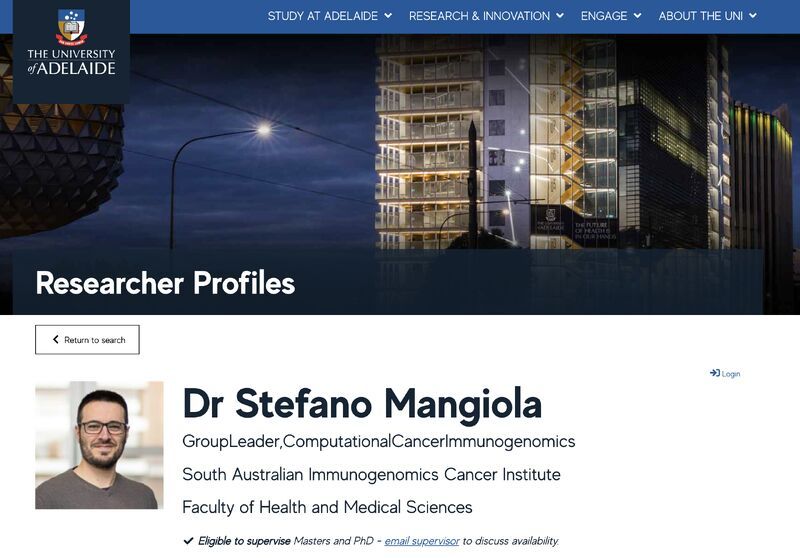
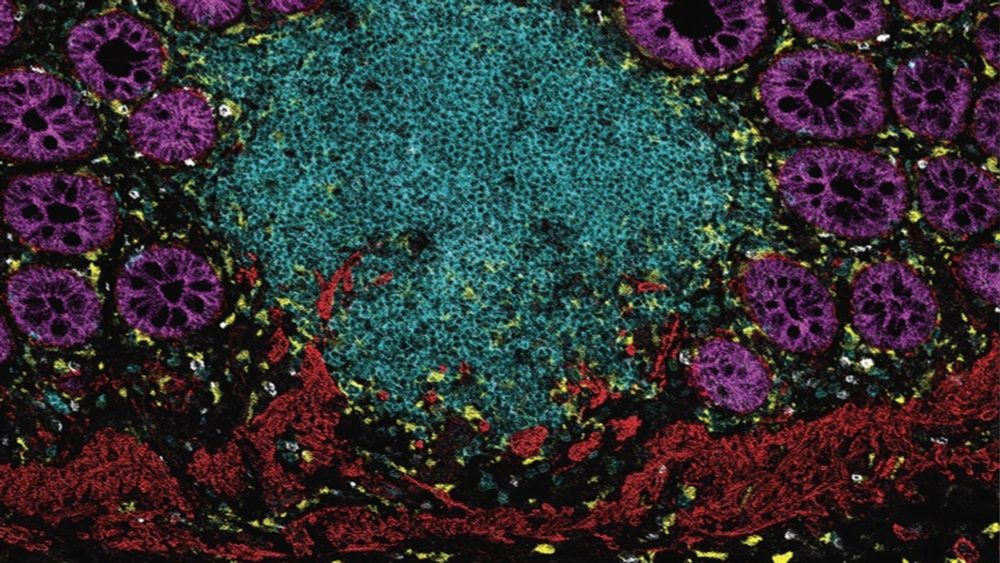
Computational cancer-immunology and tumour microenvironment
@SAiGENCI and @WEHI_research, VCA fellow @VicGovDH
Posts
Media
Videos
Starter Packs
Stefano Mangiola
@stemang.bsky.social
· Aug 26
Reposted by Stefano Mangiola
Stefano Mangiola
@stemang.bsky.social
· May 23
Stefano Mangiola
@stemang.bsky.social
· May 19
Stefano Mangiola
@stemang.bsky.social
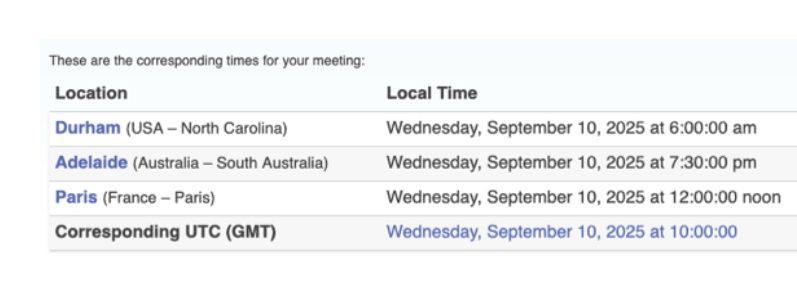
· Mar 17
Stefano Mangiola
@stemang.bsky.social
· Mar 11
Stefano Mangiola
@stemang.bsky.social
· Mar 11
Reposted by Stefano Mangiola
Stefano Mangiola
@stemang.bsky.social
· Jan 29
Stefano Mangiola
@stemang.bsky.social
· Jan 22
Stefano Mangiola
@stemang.bsky.social
· Jan 8