Sternberg Lab
@sternberglab.bsky.social
580 followers
630 following
48 posts
News from the Sternberg lab at Columbia University, Howard Hughes Medical Institute.
Posts are from lab members and not Samuel Sternberg unless signed SHS. Posts represent personal views only.
Visit us at www.sternberglab.org
Posts
Media
Videos
Starter Packs
Sternberg Lab
@sternberglab.bsky.social
· Jul 24
Sternberg Lab
@sternberglab.bsky.social
· Jul 24
Sternberg Lab
@sternberglab.bsky.social
· Jul 24
Sternberg Lab
@sternberglab.bsky.social
· Jul 24
Sternberg Lab
@sternberglab.bsky.social
· Jul 24
Sternberg Lab
@sternberglab.bsky.social
· Jul 24
Reposted by Sternberg Lab
Broad Institute
@broadinstitute.org
· Jun 24

Q&A: One scientist’s bold vision to make on-demand treatments routine for life-threatening rare genetic diseases
Broad core member and gene editing pioneer David Liu describes a framework that could enable the treatment of 1,000 patients with personalized gene editing therapies by 2030.
www.broadinstitute.org
Reposted by Sternberg Lab
Reposted by Sternberg Lab
Reposted by Sternberg Lab
Sternberg Lab
@sternberglab.bsky.social
· Jun 11
Sternberg Lab
@sternberglab.bsky.social
· Jun 11
Sternberg Lab
@sternberglab.bsky.social
· Jun 11

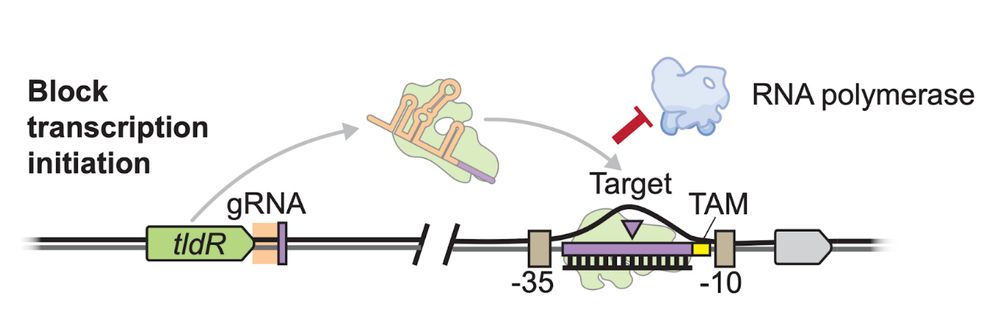
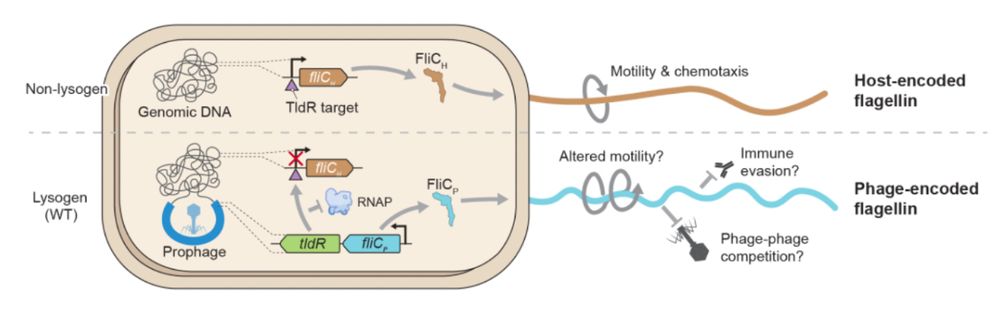
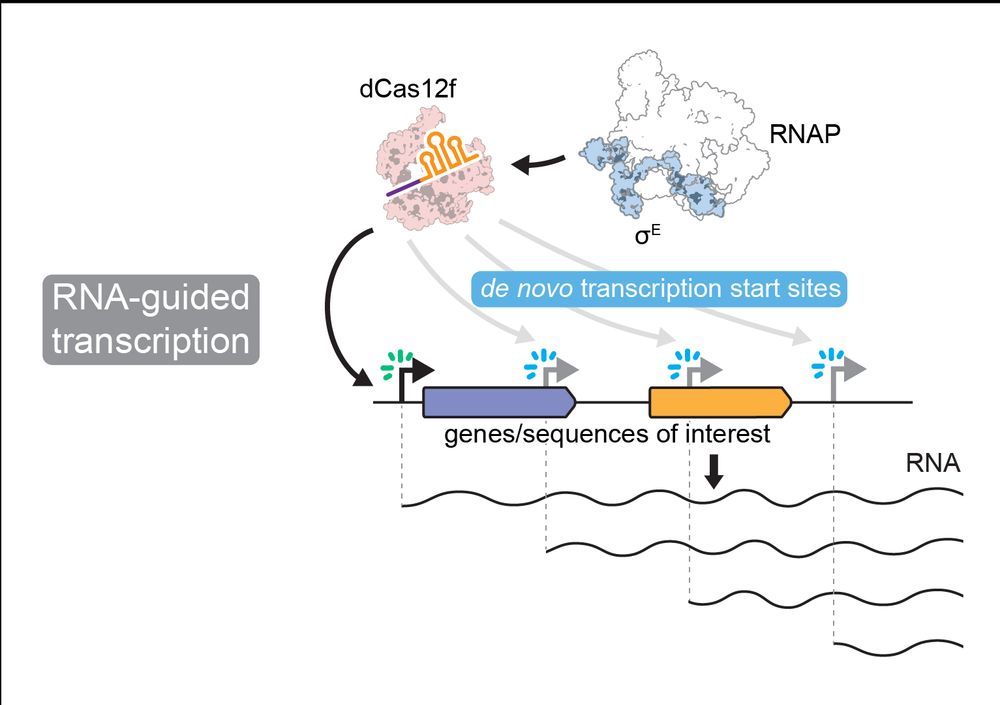
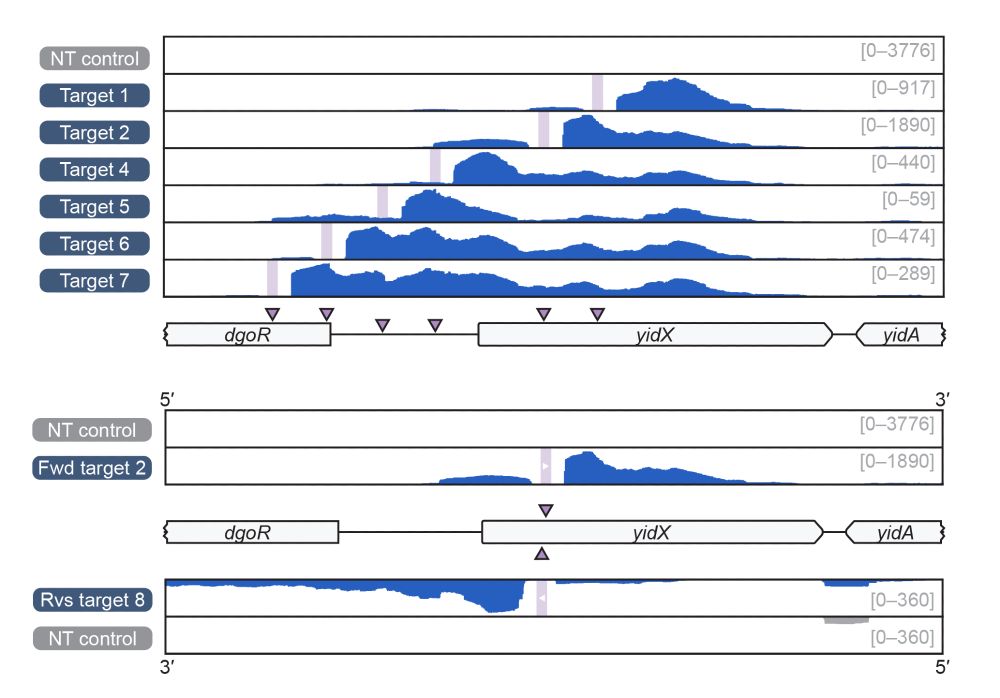
Structural basis of RNA-guided transcription by a dCas12f-σE-RNAP complex
RNA-guided proteins have emerged as critical transcriptional regulators in both natural and engineered biological systems by modulating RNA polymerase (RNAP) and its associated factors(1-5). In bacter...
www.biorxiv.org