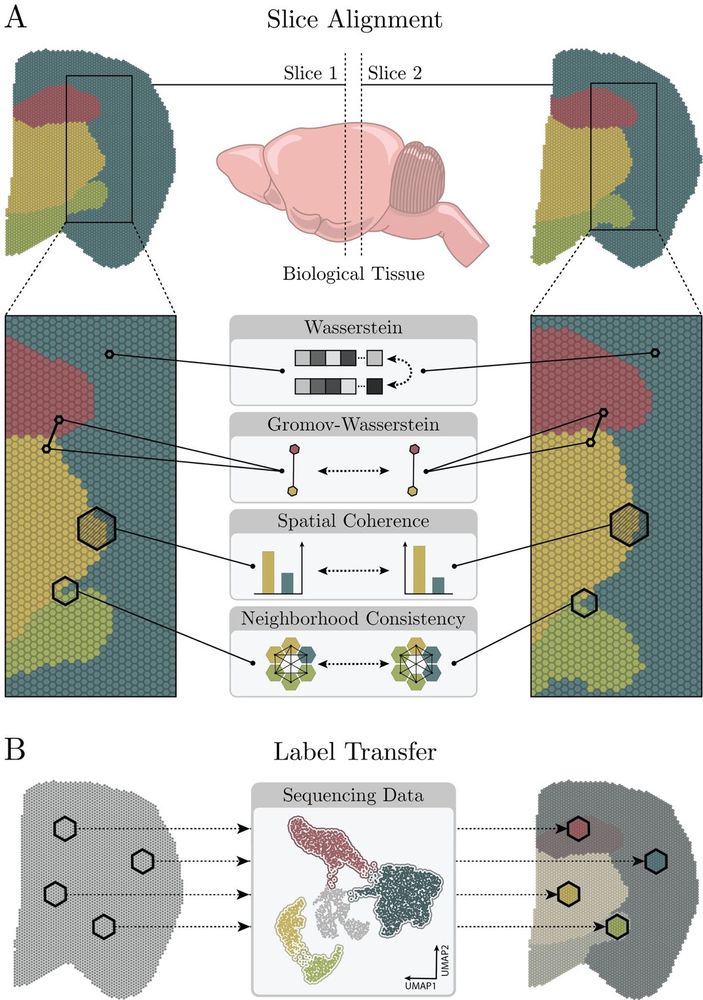
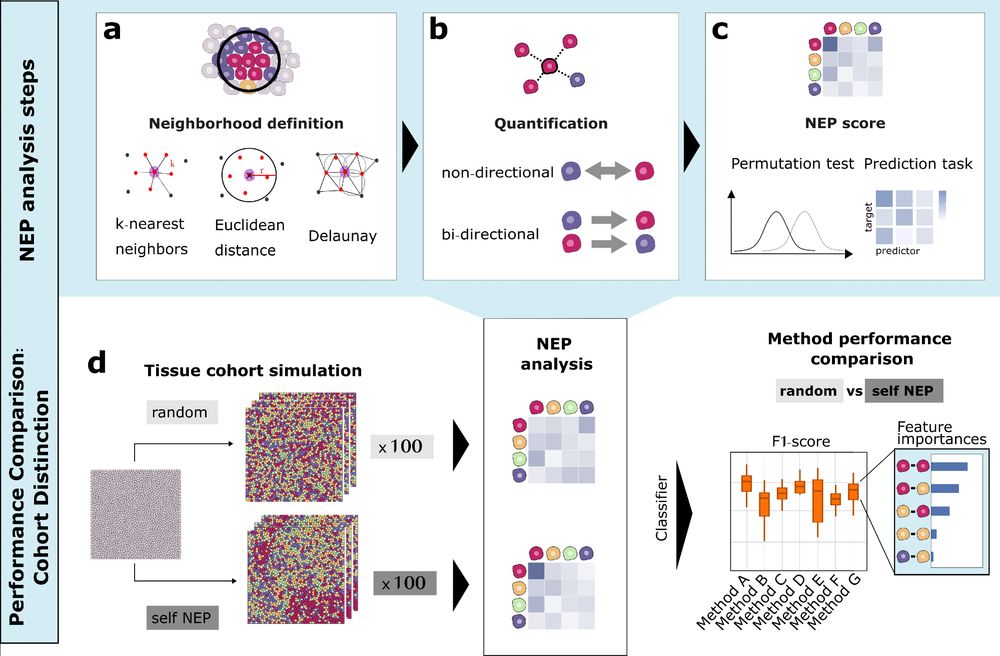
Jovan Tanevski
@tanevski.bsky.social
150 followers
91 following
7 posts
Group leader - computational biomedical discovery. Heidelberg University & Heidelberg University Hospital. https://www.tanevskilab.org
computational scientific discovery, biomedicine, spatial omics
Posts
Media
Videos
Starter Packs
Reposted by Jovan Tanevski
Reposted by Jovan Tanevski
Jovan Tanevski
@tanevski.bsky.social
· Apr 22
Jovan Tanevski
@tanevski.bsky.social
· Apr 22
Jovan Tanevski
@tanevski.bsky.social
· Apr 15