Tristan Bepler
@tbepler.bsky.social
400 followers
390 following
32 posts
Scientist and Group Leader of the Simons Machine Learning Center
@SEMC_NYSBC. Co-founder and CEO of http://OpenProtein.AI. Opinions are my own.
Posts
Media
Videos
Starter Packs
Pinned
Tristan Bepler
@tbepler.bsky.social
· Feb 11
Tristan Bepler
@tbepler.bsky.social
· Aug 26

Understanding protein function with a multimodal retrieval-augmented foundation model
Protein language models (PLMs) learn probability distributions over natural protein sequences. By learning from hundreds of millions of natural protein sequences, protein understanding and design capa...
arxiv.org
Reposted by Tristan Bepler
Reposted by Tristan Bepler
Tristan Bepler
@tbepler.bsky.social
· Jun 20
Tristan Bepler
@tbepler.bsky.social
· Jun 20
Reposted by Tristan Bepler
Pascal Notin
@pascalnotin.bsky.social
· May 8
Tristan Bepler
@tbepler.bsky.social
· May 12
Tristan Bepler
@tbepler.bsky.social
· May 12
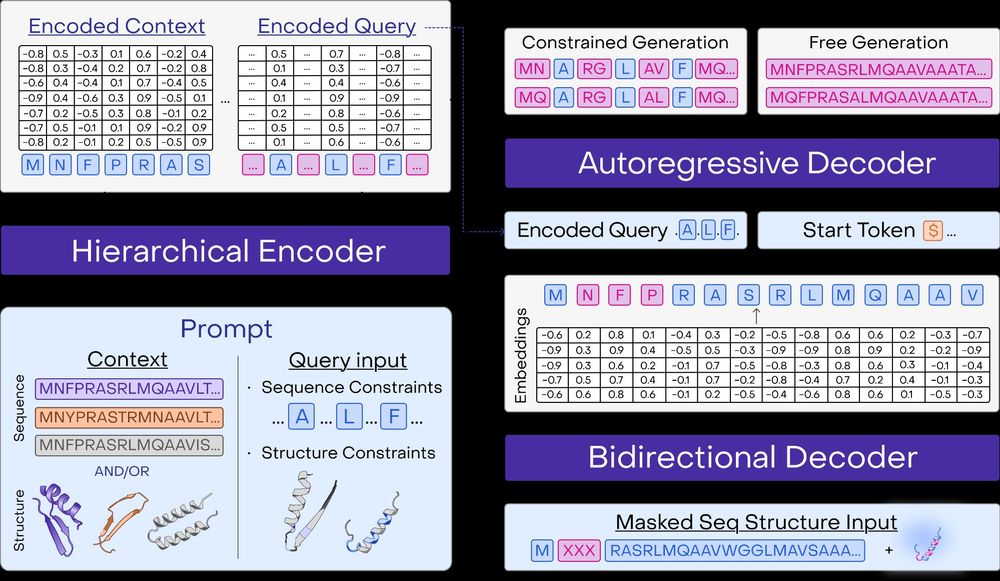
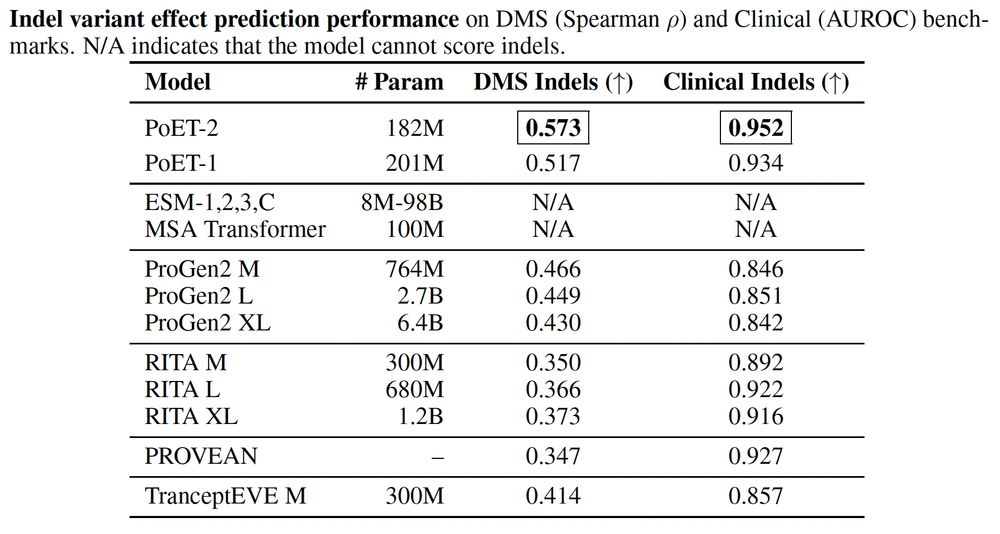
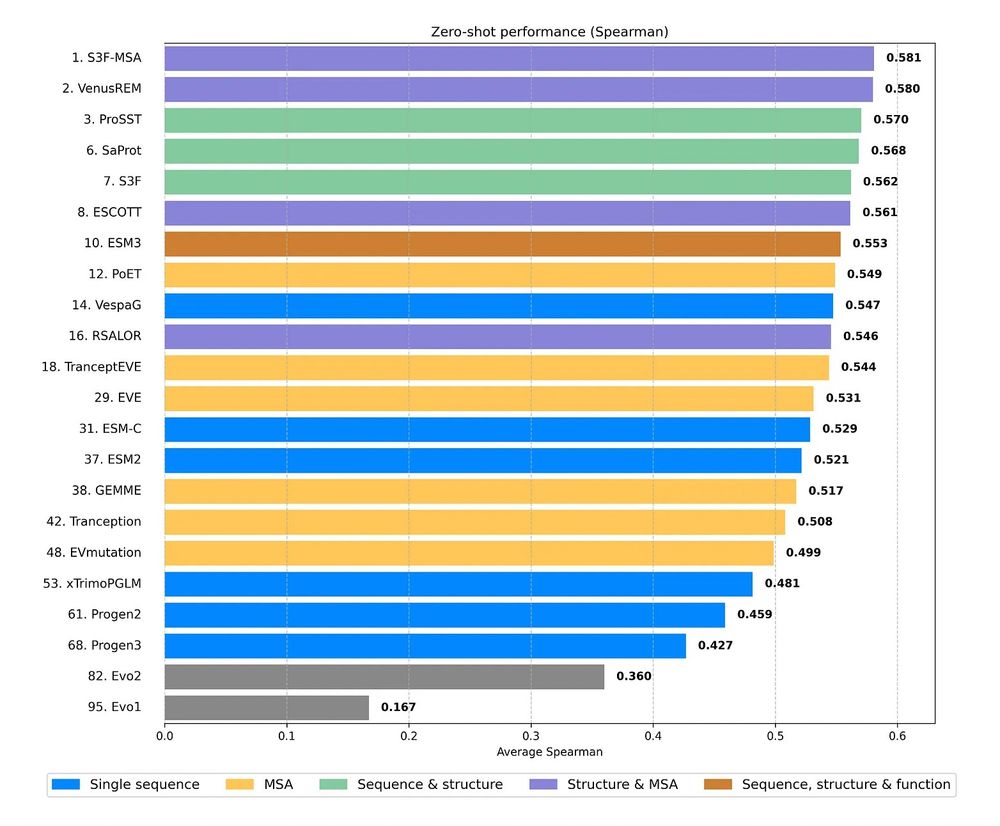
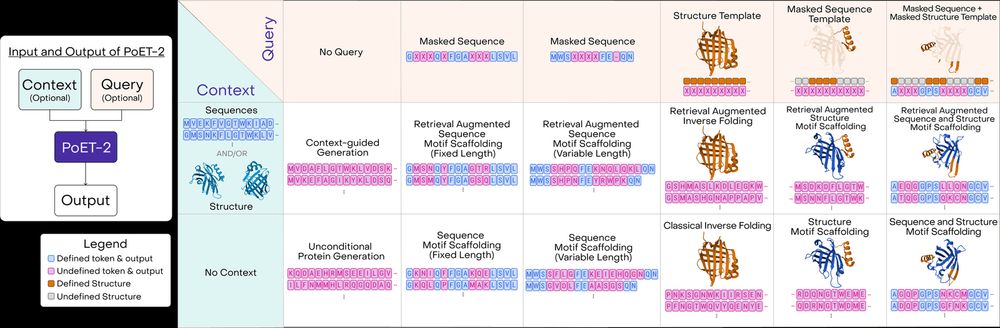
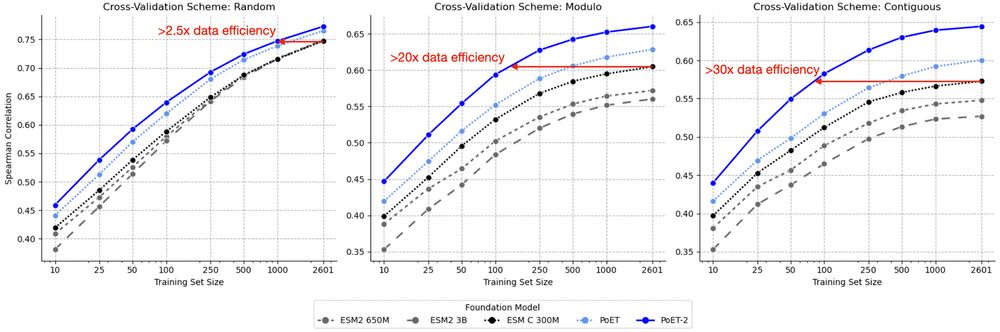
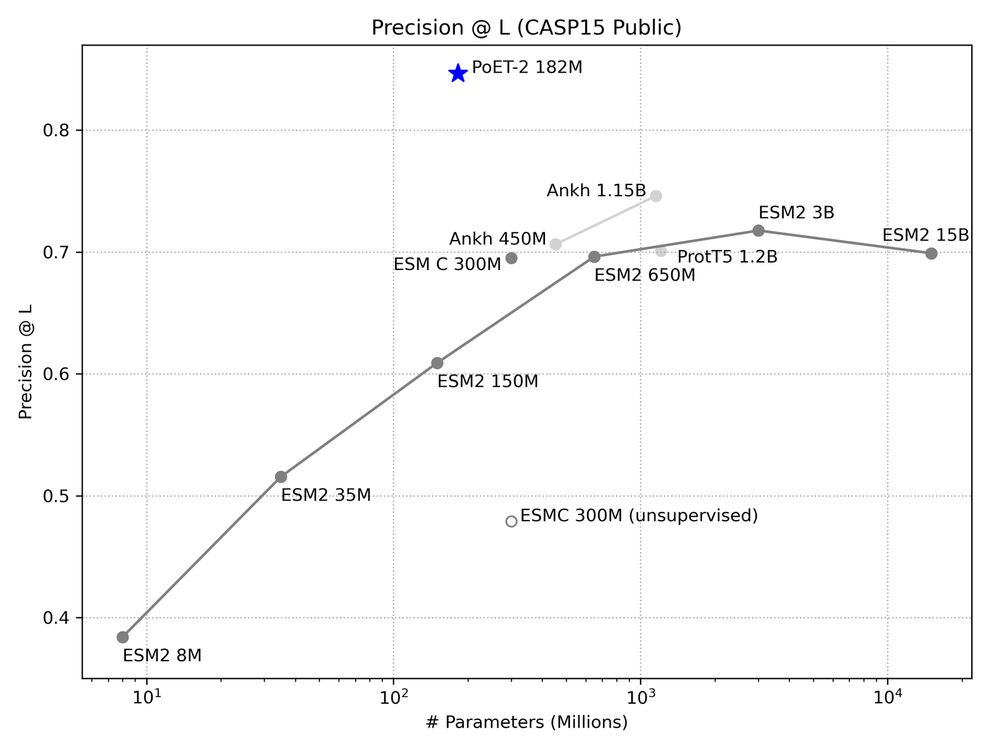
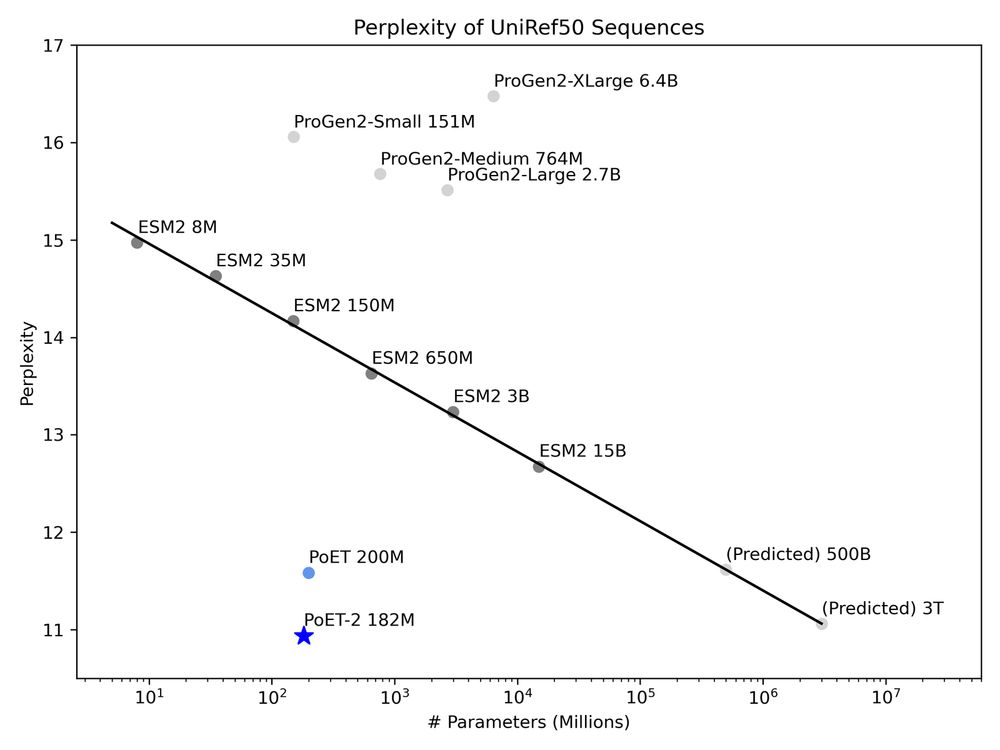
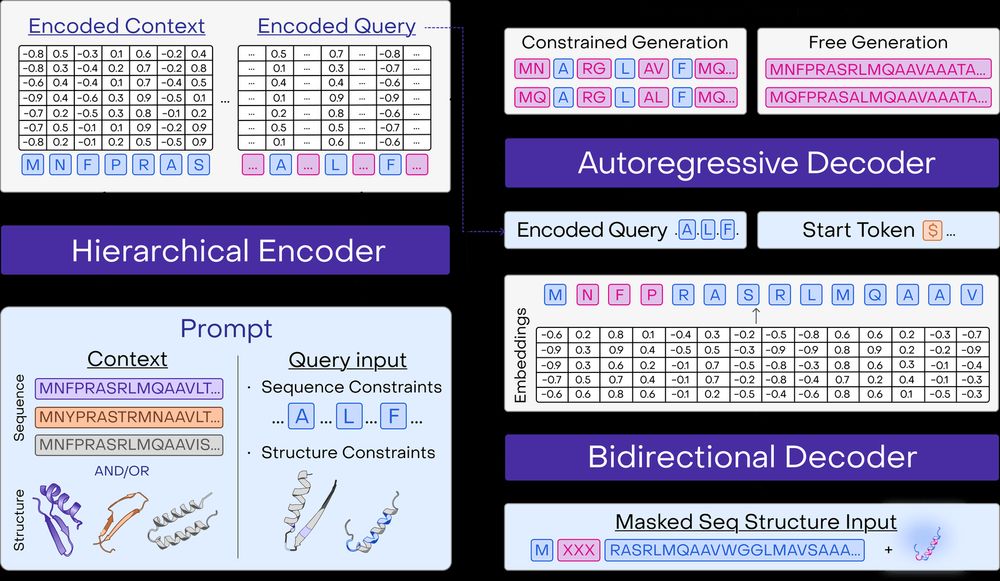
A multimodal foundation model for controllable protein generation and representation learning
PoET-2 is a next generation protein language model that transforms our ability to engineer proteins by learning from nature’s design principles. Through its unique multimodal architecture and training...
www.openprotein.ai
Reposted by Tristan Bepler
Tristan Bepler
@tbepler.bsky.social
· Feb 11
Tristan Bepler
@tbepler.bsky.social
· Feb 11
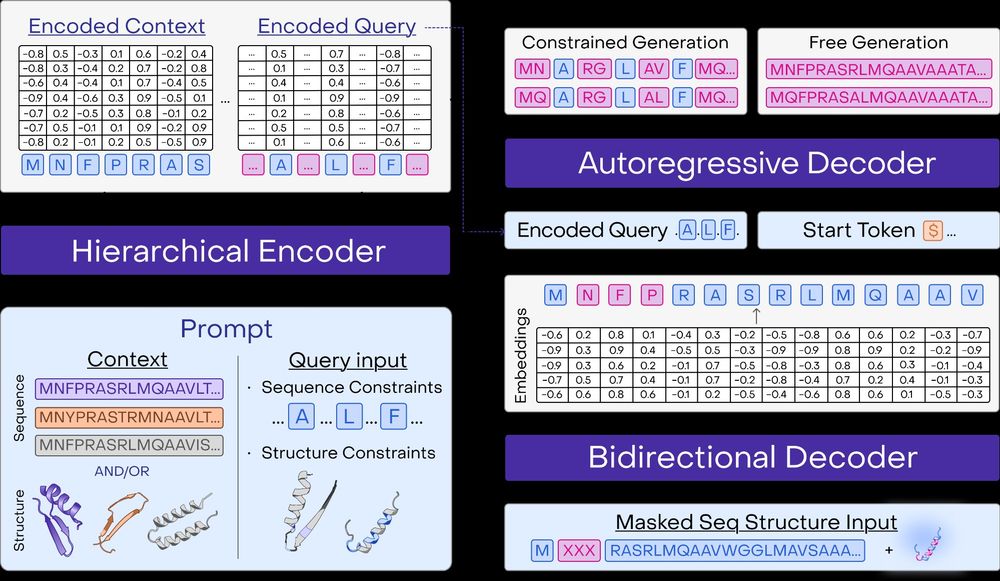
A multimodal foundation model for controllable protein generation and representation learning
PoET-2 is a next generation protein language model that transforms our ability to engineer proteins by learning from nature’s design principles. Through its unique multimodal architecture and training...
www.openprotein.ai
Tristan Bepler
@tbepler.bsky.social
· Feb 11
Tristan Bepler
@tbepler.bsky.social
· Feb 11
Tristan Bepler
@tbepler.bsky.social
· Feb 11
Tristan Bepler
@tbepler.bsky.social
· Feb 11