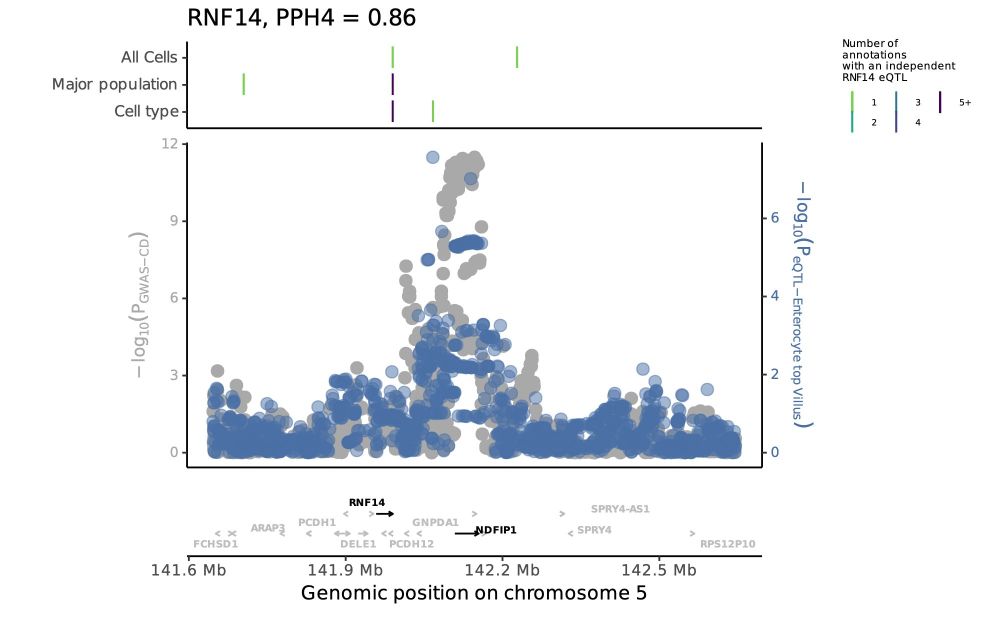
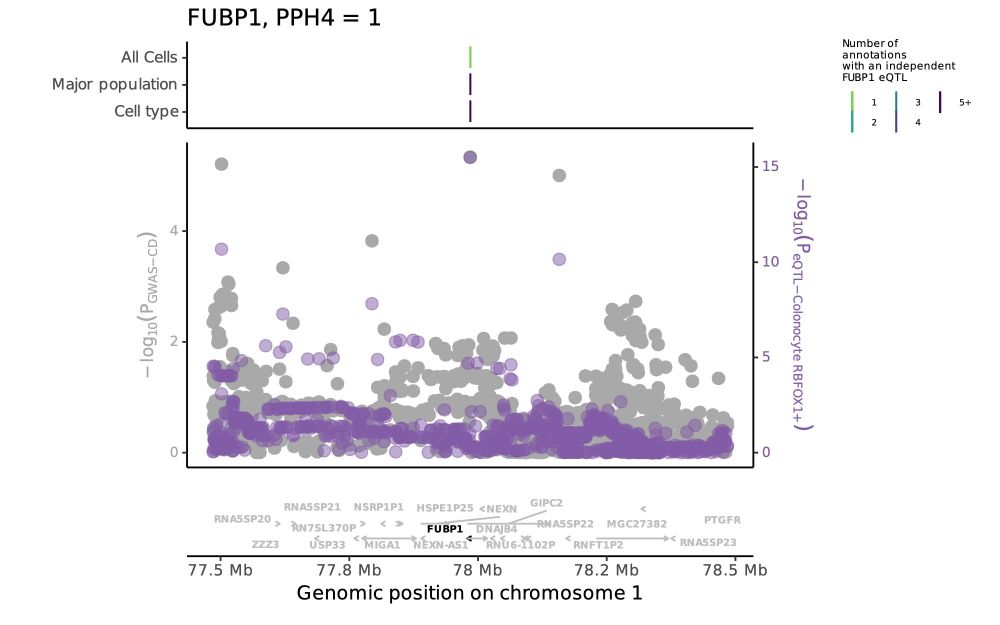
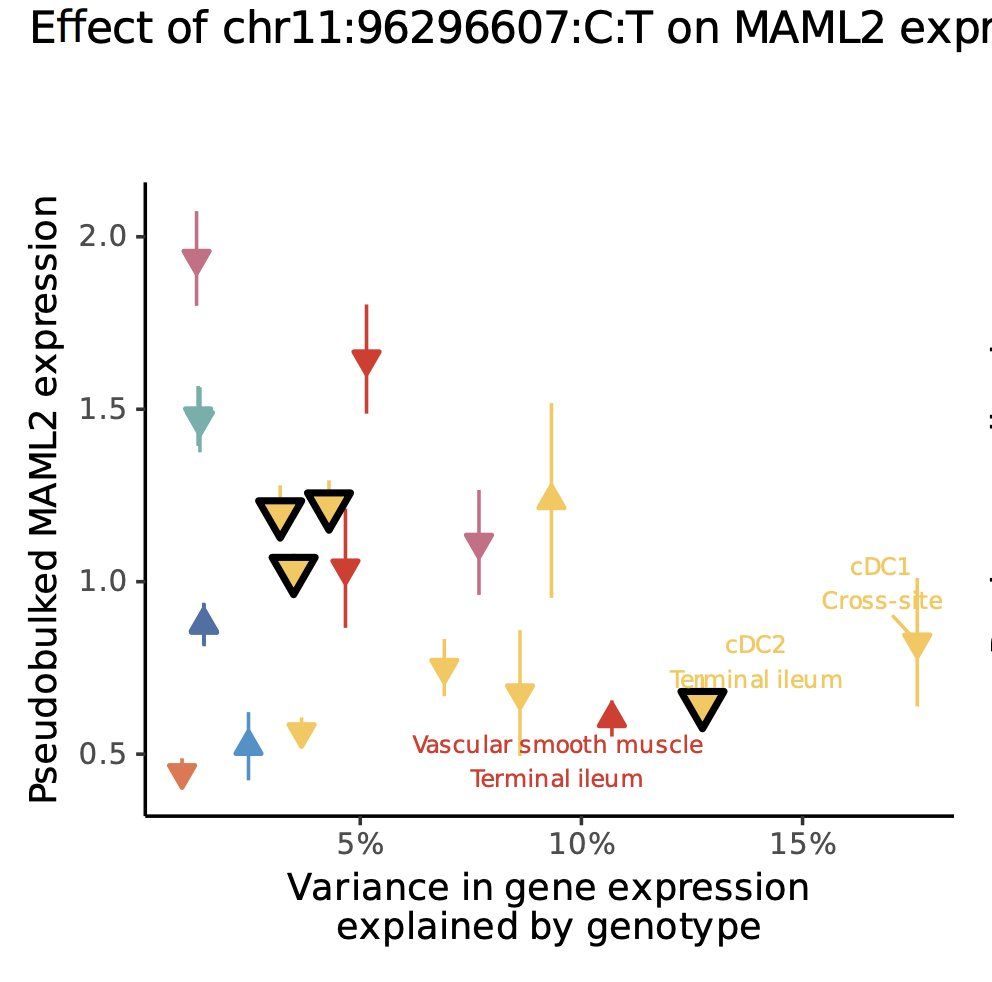
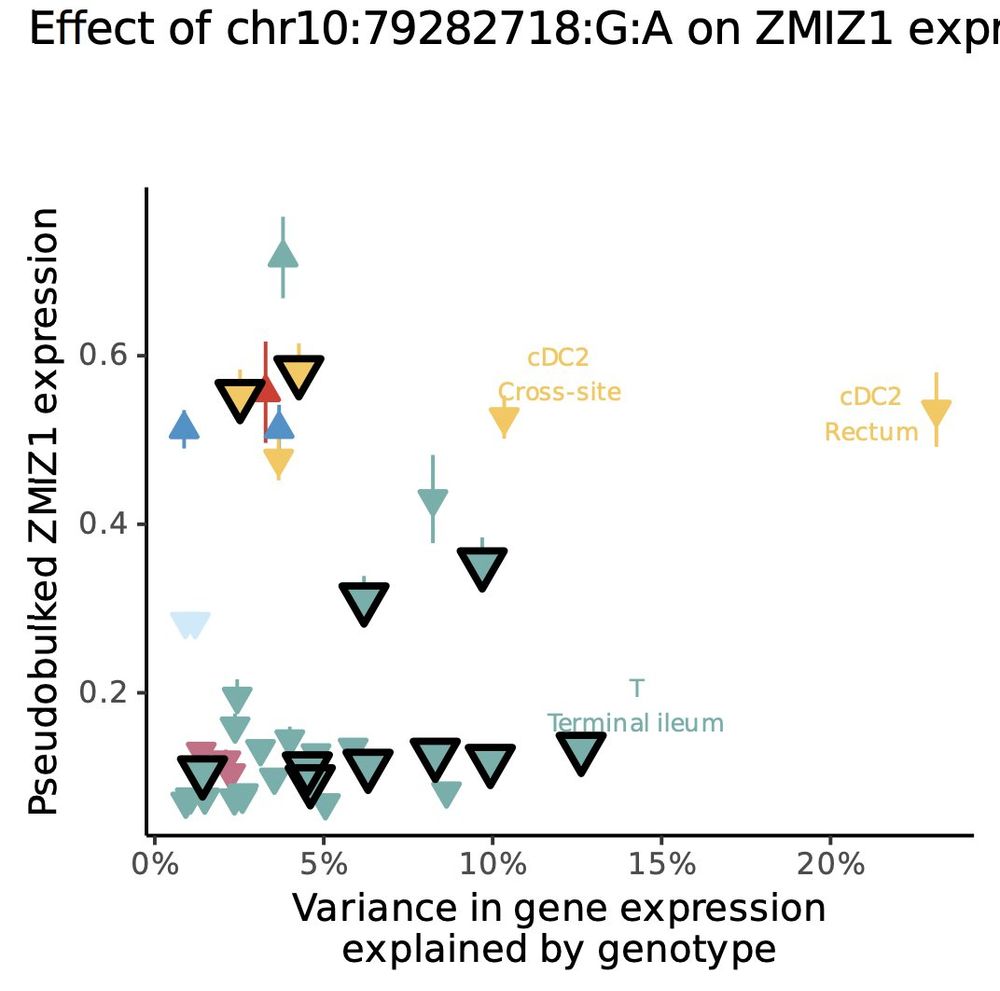
Tobi Alegbe
@tobioinformatics.bsky.social
120 followers
170 following
28 posts
I look at small things on a big scale
Single-cell data scientist at Open Targets, EMBL-EBI. Previously StatGen PhD student at Sanger Institute
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Tobi Alegbe