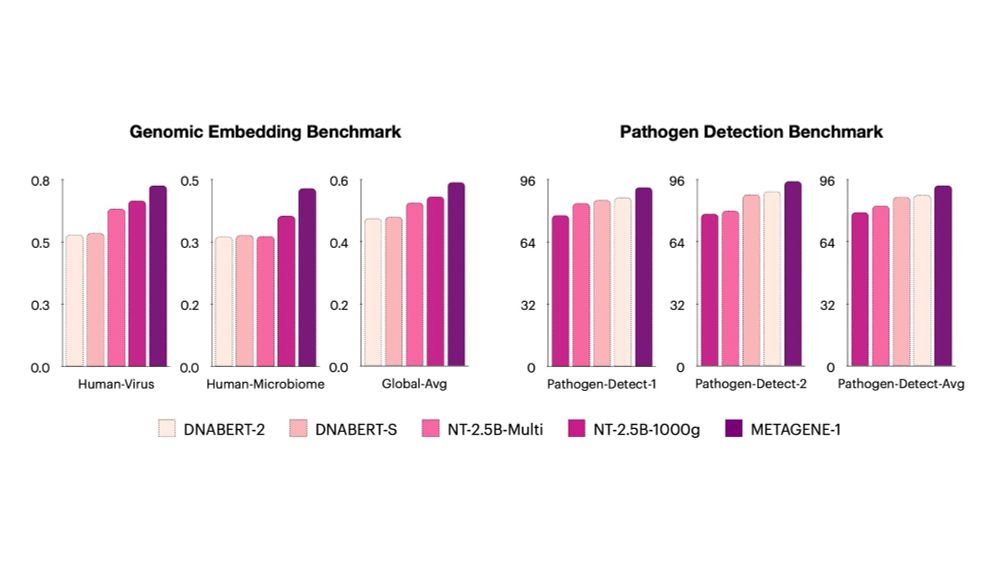
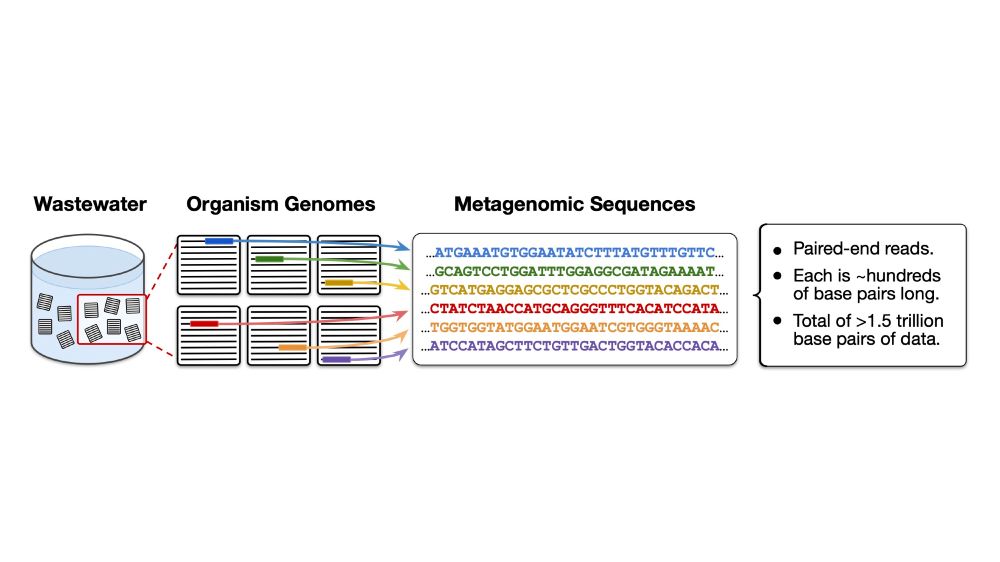
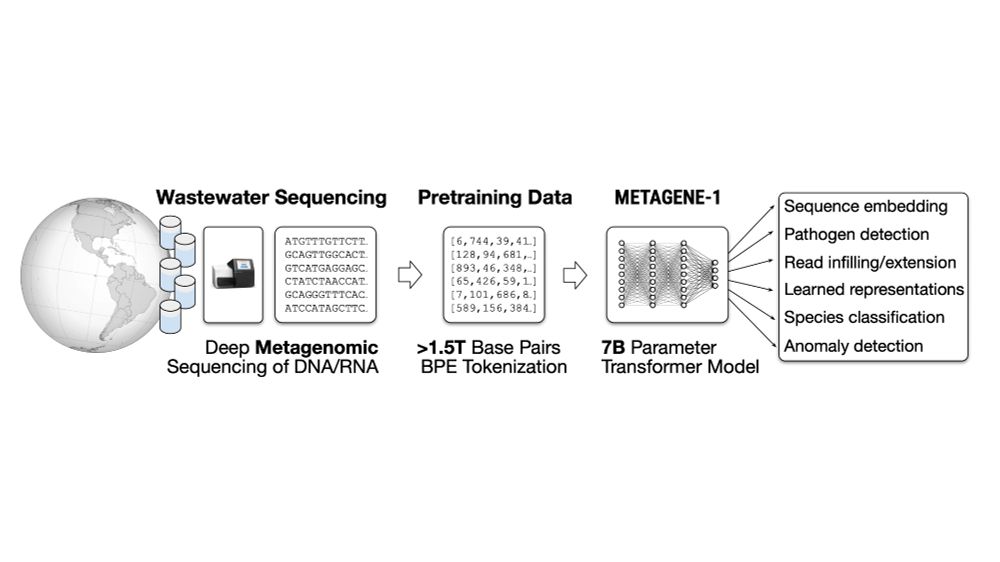
Willie Neiswanger
@willieneis.bsky.social
7K followers
190 following
240 posts
Assistant Professor in CS + AI at USC. Previously at Stanford, CMU. Machine Learning, Decision Making, AI-for-Science, Generative AI, ML Systems, LLMs.
https://willieneis.github.io
Posts
Media
Videos
Starter Packs
Pinned
Willie Neiswanger
@willieneis.bsky.social
· Nov 10
Reposted by Willie Neiswanger
Reposted by Willie Neiswanger
Willie Neiswanger
@willieneis.bsky.social
· Feb 12
Reposted by Willie Neiswanger
Reposted by Willie Neiswanger
Reposted by Willie Neiswanger
Willie Neiswanger
@willieneis.bsky.social
· Nov 30
Willie Neiswanger
@willieneis.bsky.social
· Nov 30
Willie Neiswanger
@willieneis.bsky.social
· Nov 28
Willie Neiswanger
@willieneis.bsky.social
· Nov 28