Aino-Maija Leppä
@amleppa.bsky.social
110 followers
60 following
5 posts
Current | Postdoc at Francis Crick Institute with Charles Swanton exploring lung cancer evolution.
Before | PhD at DKFZ with Andreas Trumpp charting AML heterogeneity.
Posts
Media
Videos
Starter Packs
Reposted by Aino-Maija Leppä
The Francis Crick Institute
@crick.ac.uk
· Mar 11
Beneficial genetic changes observed in regular blood donors
Researchers at the Francis Crick Institute have identified genetic changes in blood stem cells from frequent blood donors that support the production of new, non-cancerous cells.
www.crick.ac.uk
Reposted by Aino-Maija Leppä
Reposted by Aino-Maija Leppä
Aino-Maija Leppä
@amleppa.bsky.social
· Nov 25
Aino-Maija Leppä
@amleppa.bsky.social
· Nov 25

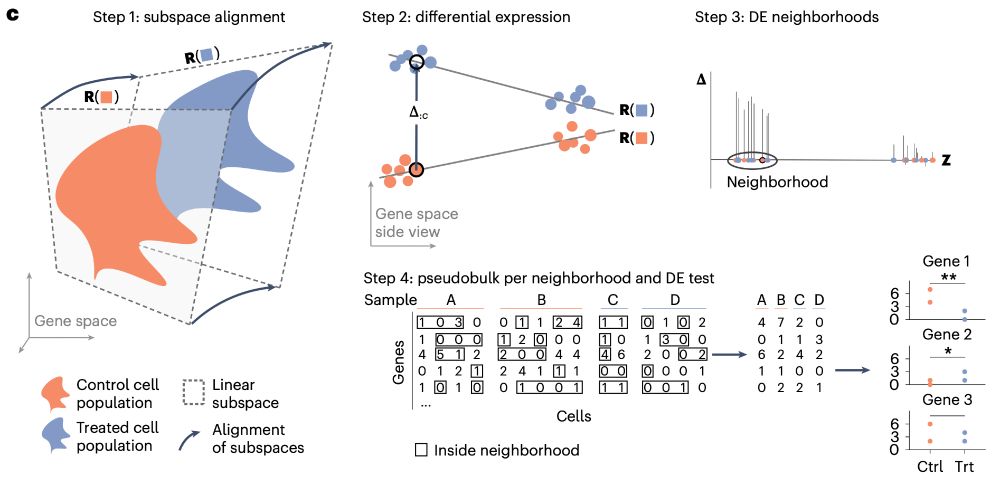
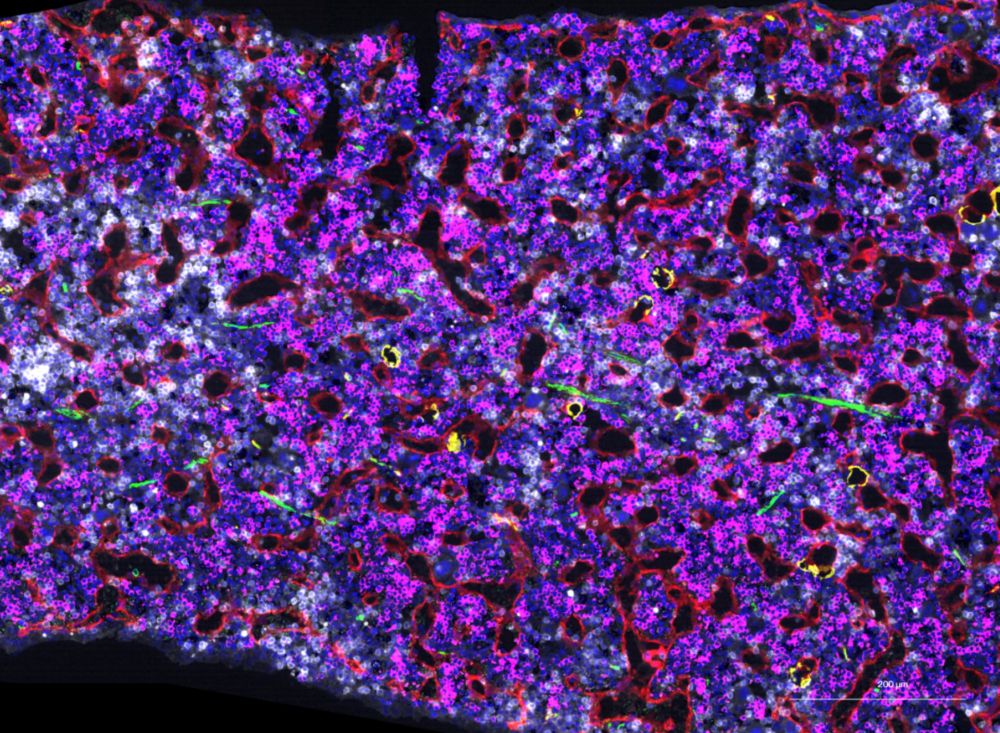
Single-cell multiomics analysis reveals dynamic clonal evolution and targetable phenotypes in acute myeloid leukemia with complex karyotype - Nature Genetics
An integrated single-cell multiomic analysis of complex karyotype acute myeloid leukemia characterizes intratumoral heterogeneity and highlights links to therapeutic sensitivities.
www.nature.com