Roberto Chica Lab
@chicalab.bsky.social
1.5K followers
200 following
38 posts
Our research group at the University of Ottawa specializes in computational enzyme design.
mysite.science.uottawa.ca/rchica/
Posts
Media
Videos
Starter Packs
Roberto Chica Lab
@chicalab.bsky.social
· Aug 26

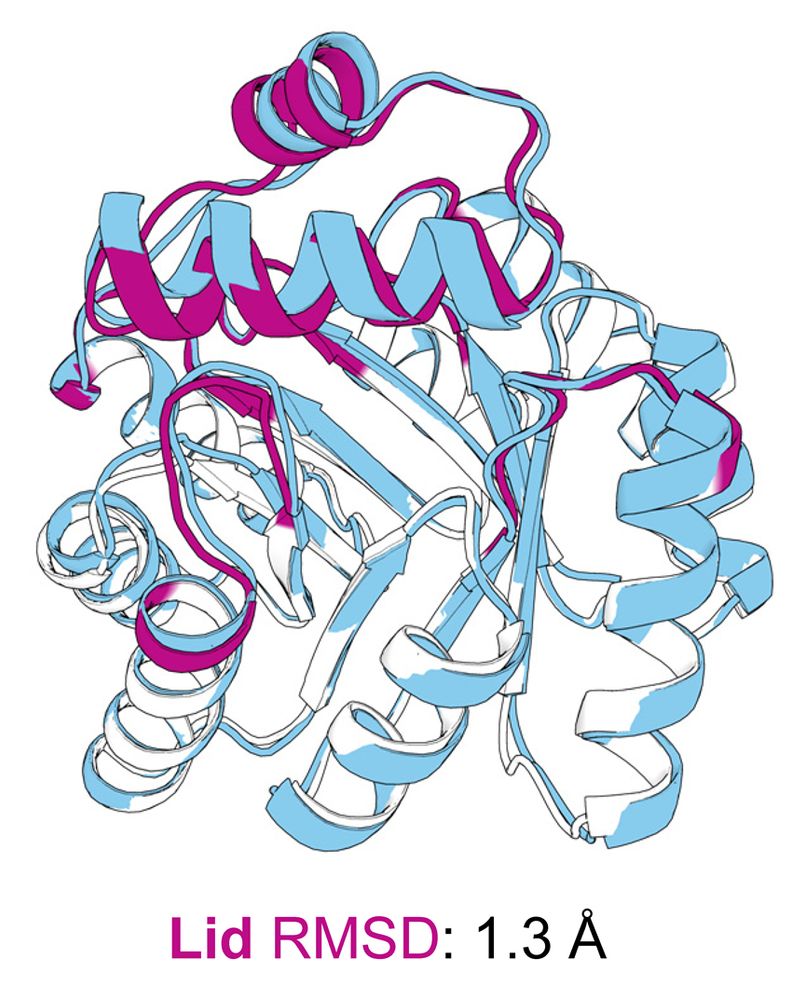
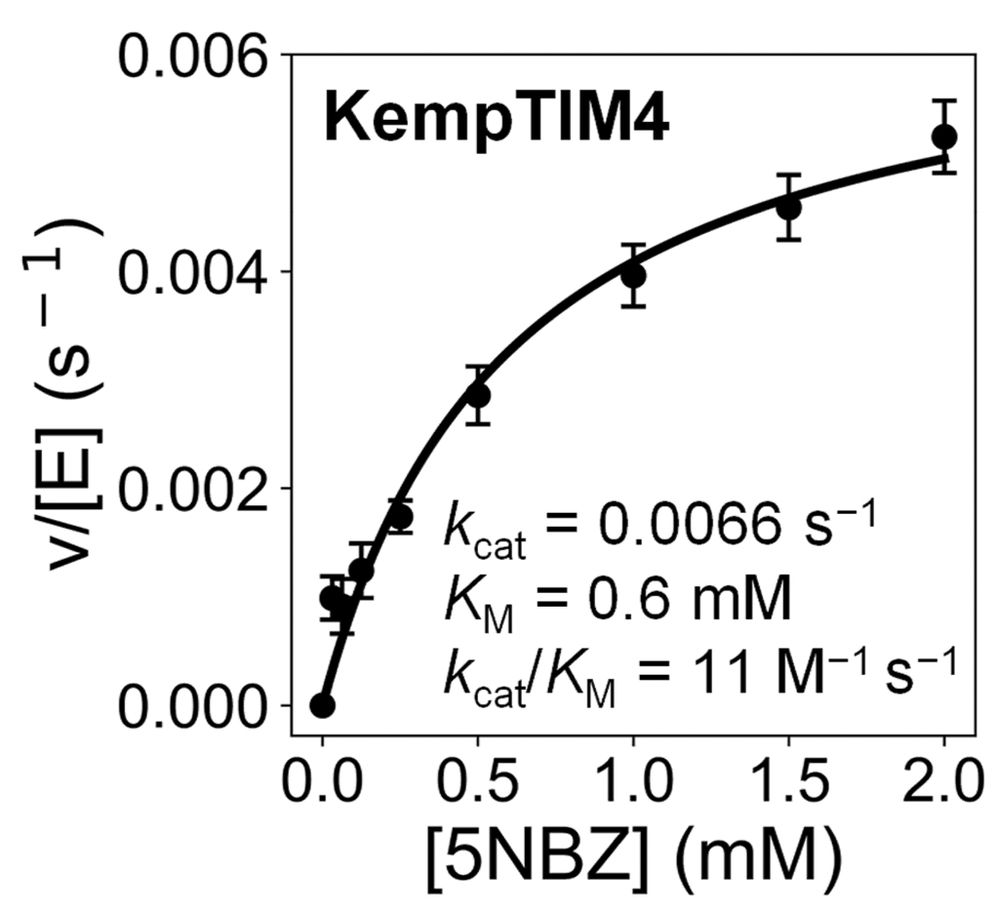
Distal Mutations in a Designed Retro-Aldolase Alter Loop Dynamics to Shift and Accelerate the Rate-Limiting Step
Amino acid residues distant from an enzyme’s active site are known to influence catalysis, but their mechanistic contributions to the catalytic cycle remain poorly understood. Here, we investigate the...
pubs.acs.org
Roberto Chica Lab
@chicalab.bsky.social
· Jul 31
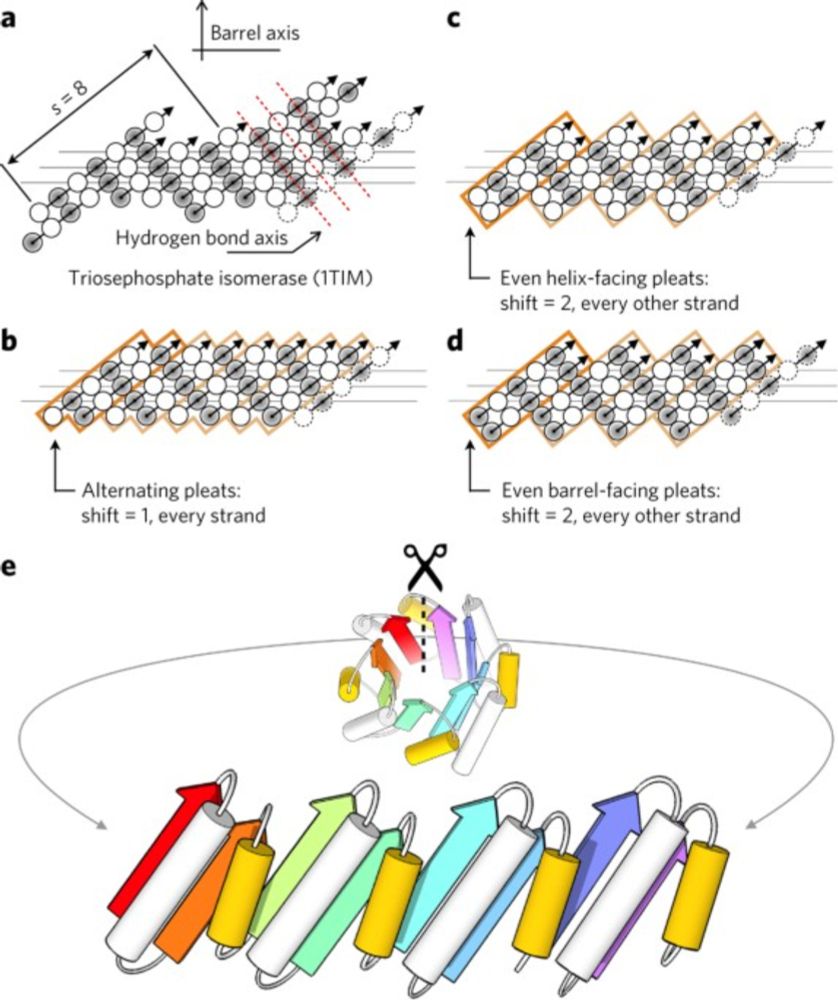
De novo design of a four-fold symmetric TIM-barrel protein with atomic-level accuracy - Nature Chemical Biology
Despite substantial effort, the de novo design of a stable TIM-barrel protein fold has remained elusive. A Rosetta-based computational strategy identifies a unique 184-residue sequence that adopts a T...
www.nature.com
Roberto Chica Lab
@chicalab.bsky.social
· Jul 31

Characterization of a Helical Protein Designed from First Principles
The question of how the primary amino acid sequence of a protein determines its three-dimensional structure is still unanswered. One approach to this problem involves the de novo design of model pepti...
www.science.org
Roberto Chica Lab
@chicalab.bsky.social
· Jul 29
Roberto Chica Lab
@chicalab.bsky.social
· Jul 29

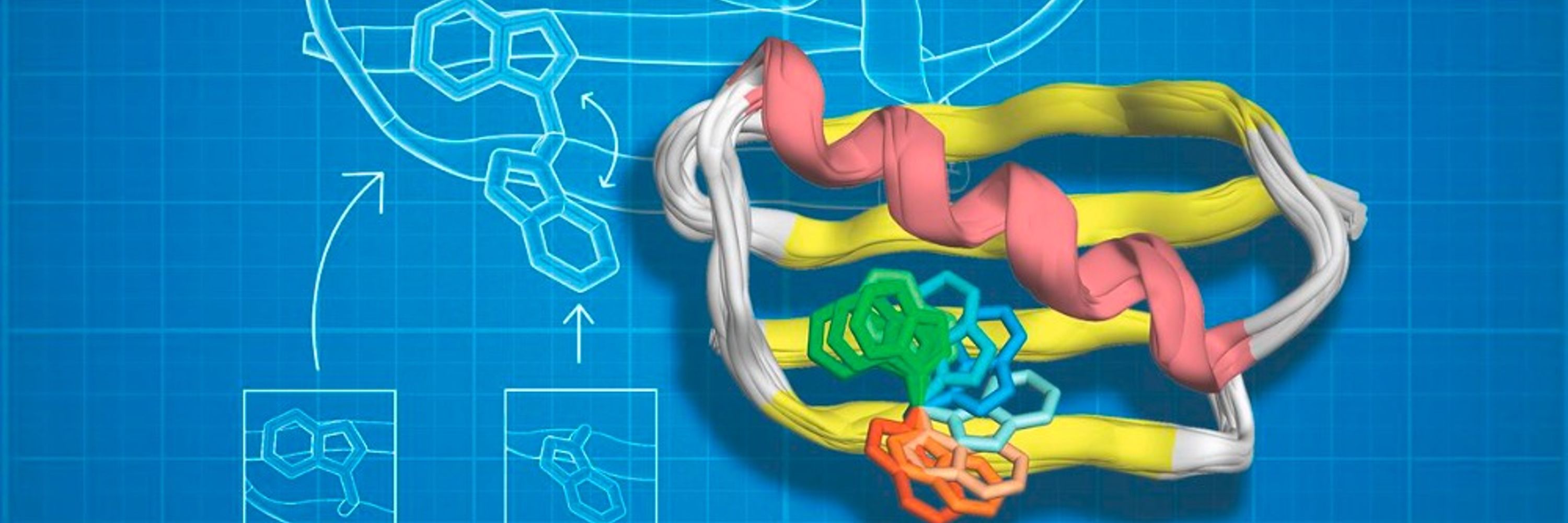
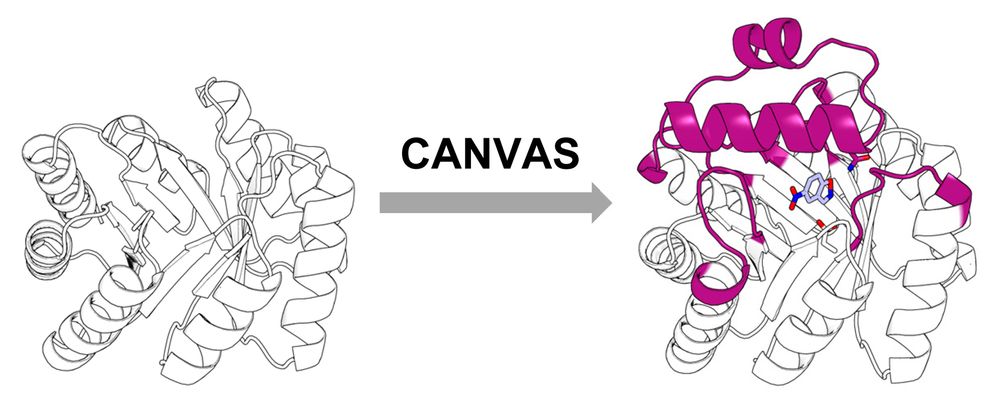
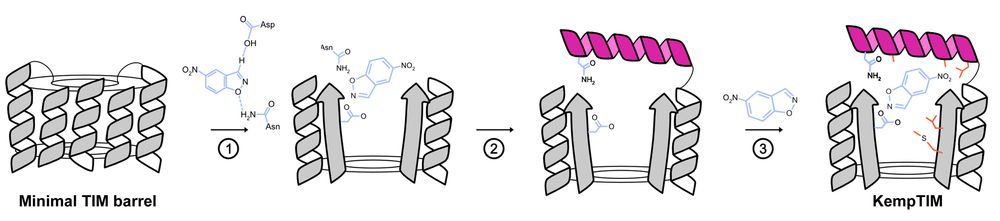
Customizing the Structure of a Minimal TIM Barrel to Craft a De Novo Enzyme
The TIM barrel is the most prevalent fold in natural enzymes, supporting efficient catalysis of diverse chemical reactions. While de novo TIM barrels have been successfully designed, their minimalisti...
www.biorxiv.org
Roberto Chica Lab
@chicalab.bsky.social
· Jul 25
Reposted by Roberto Chica Lab
Reposted by Roberto Chica Lab
Nick Polizzi
@nickpolizzi.bsky.social
· Apr 28

Zero-shot design of drug-binding proteins via neural selection-expansion
Computational design of molecular recognition remains challenging despite advances in deep learning. The design of proteins that bind to small molecules has been particularly difficult because it requ...
www.biorxiv.org
Reposted by Roberto Chica Lab
Roberto Chica Lab
@chicalab.bsky.social
· Feb 28

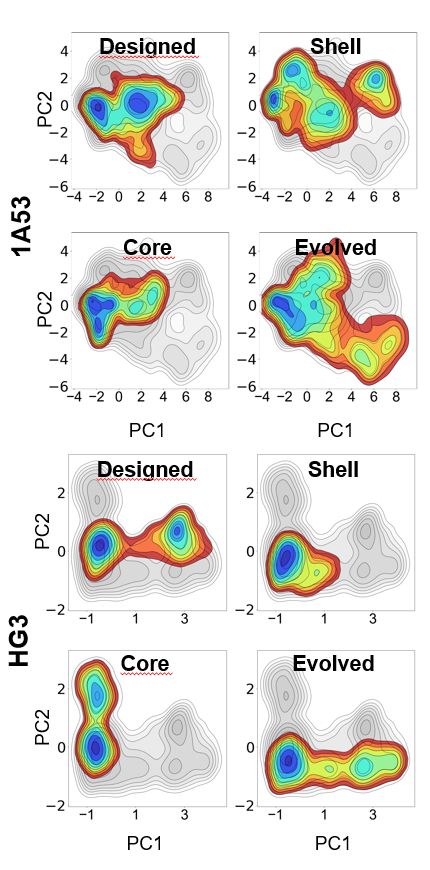
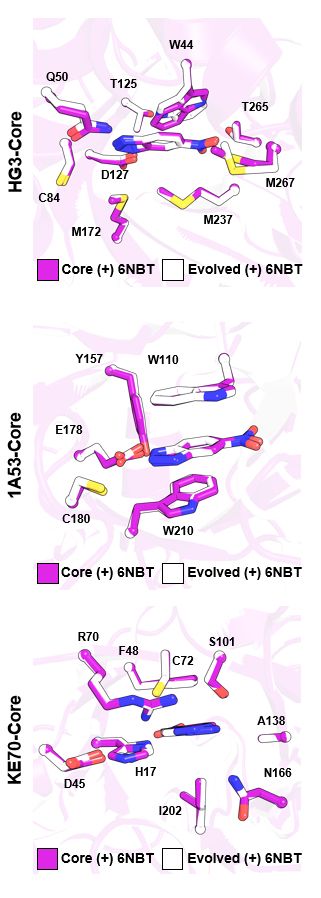
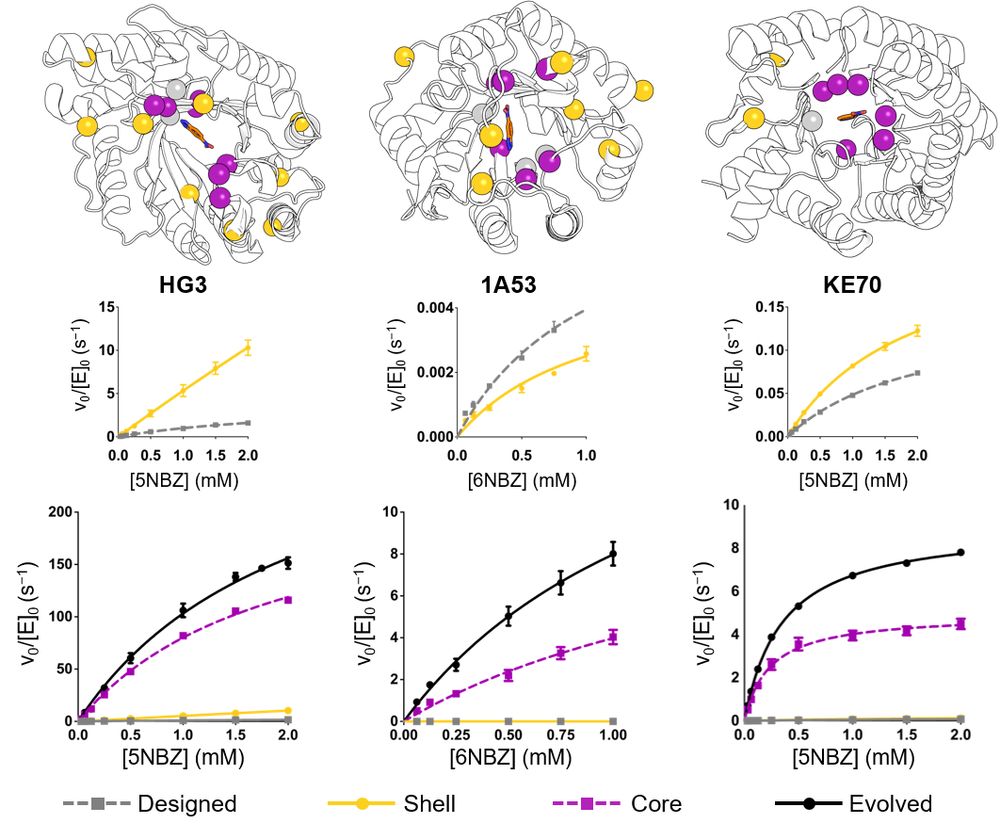
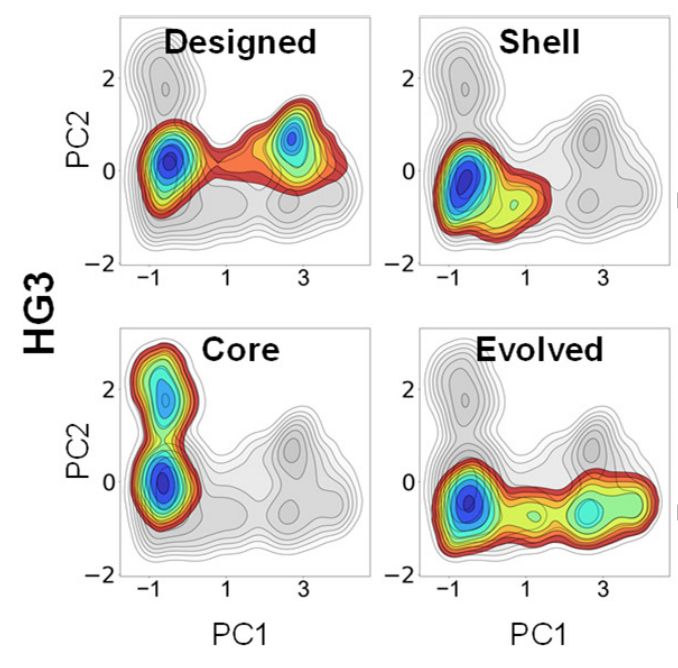
Distal mutations enhance catalysis in designed enzymes by facilitating substrate binding and product release
The role of amino-acid residues distant from an enzyme's active site in facilitating the complete catalytic cycle—including substrate binding, chemical transformation, and product release—remains poor...
www.biorxiv.org
Roberto Chica Lab
@chicalab.bsky.social
· Feb 28
Roberto Chica Lab
@chicalab.bsky.social
· Feb 28
Reposted by Roberto Chica Lab
Roberto Chica Lab
@chicalab.bsky.social
· Feb 27

Distal mutations in a designed retro-aldolase alter loop dynamics to shift and accelerate the rate-limiting step
Amino-acid residues distant from an enzyme’s active site are known to influence catalysis, but their mechanistic contributions to the catalytic cycle remain poorly understood. Here, we investigate the...
www.biorxiv.org