
Opinions my own
https://linktr.ee/ddelalamo

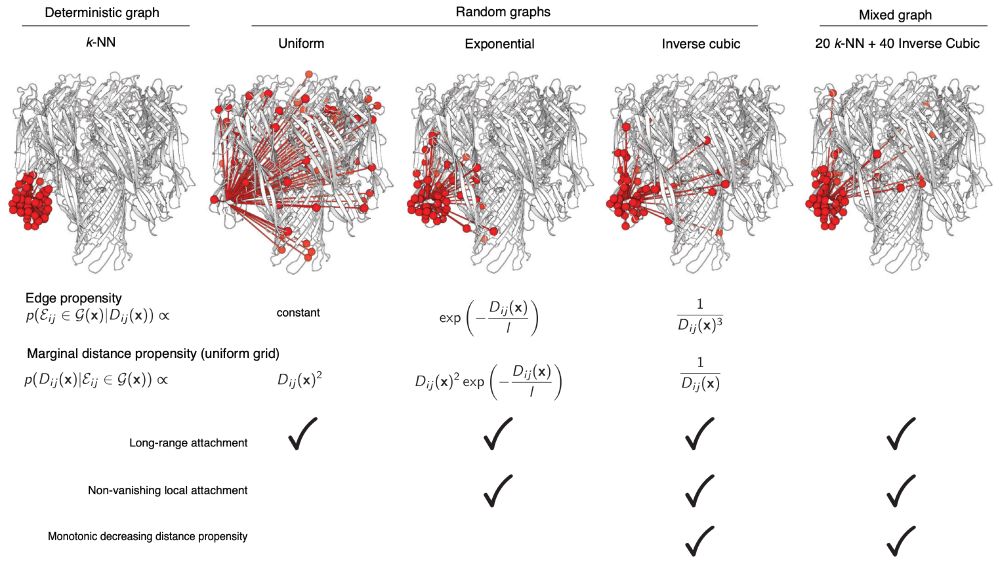

openreview.net/pdf/dd8aa97e...

openreview.net/pdf/dd8aa97e...




One of their creations is this beautiful flower-shaped tetrameric pore 🌼, a brand new member of the #ProteinCosmos 🧶🧬 🧪

One of their creations is this beautiful flower-shaped tetrameric pore 🌼, a brand new member of the #ProteinCosmos 🧶🧬 🧪



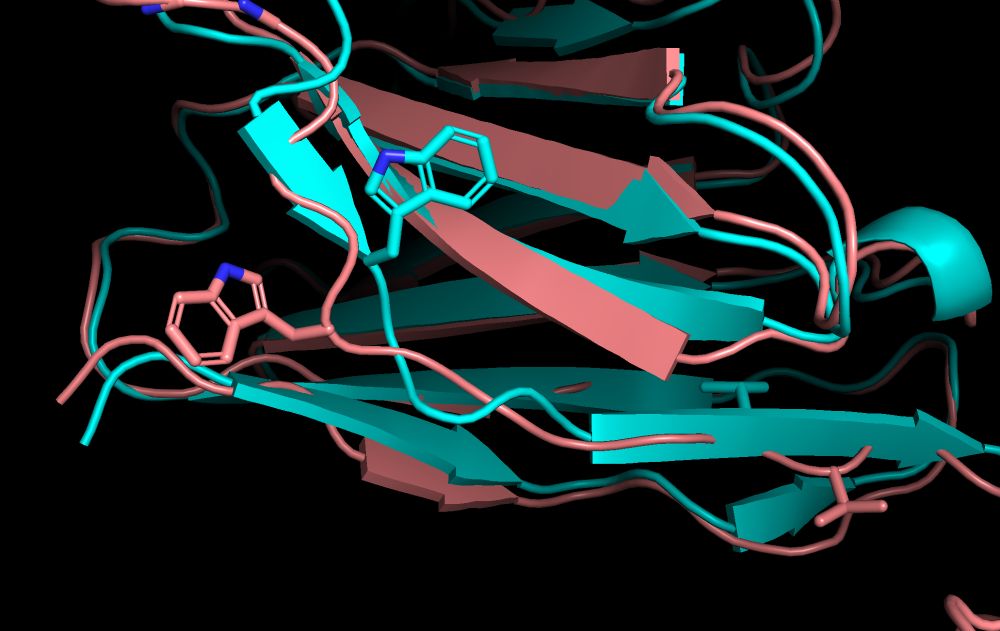

Ab design via seq/struct conditioning designs VHHs. Million-scale screens show success.






Germinal produces functional nanobodies in just dozens of tests, making custom antibody design more accessible than ever before.

jobs.careers.microsoft.com/global/en/jo...
jobs.careers.microsoft.com/global/en/jo...

1. Fill out a form online
2. Pay a processing fee, also online
3. Print form and put it in an envelope and MAIL it
4. Receive criminal record by email
1. Fill out a form online
2. Pay a processing fee, also online
3. Print form and put it in an envelope and MAIL it
4. Receive criminal record by email


