VSPGx: NGS data to pharmacogenomic diplotypes, auto-annotating w/ CPIC/FDA drug info, improving tools.

VSPGx: NGS data to pharmacogenomic diplotypes, auto-annotating w/ CPIC/FDA drug info, improving tools.
Analyzes pathway activity using graph-based belief propagation to handle feedback loops and integrate gene expression with topology.



Analyzes pathway activity using graph-based belief propagation to handle feedback loops and integrate gene expression with topology.
Analyzes DNA methylation sequence determinants. DL model predicts profiles across tissues & IDs assoc. motifs.




Analyzes DNA methylation sequence determinants. DL model predicts profiles across tissues & IDs assoc. motifs.
Cas9 nanopore seq: simultaneous STR expansion/methylation detection in neurogenetic disorders.




Cas9 nanopore seq: simultaneous STR expansion/methylation detection in neurogenetic disorders.
Aligns multi-omics data with frozen transformers using PEFT. Aligns cross-sample and modality data, yielding robust embeddings with batch invariance.




Aligns multi-omics data with frozen transformers using PEFT. Aligns cross-sample and modality data, yielding robust embeddings with batch invariance.
Models human proteome's disordered regions by creating conformer ensembles based on AlphaFold2 folded domains, enhancing biological relevance.




Models human proteome's disordered regions by creating conformer ensembles based on AlphaFold2 folded domains, enhancing biological relevance.
LMMs can name objects/actions but struggle to precisely detect "contact" and "release" events in videos, indicating a lack of perceptual grounding.




LMMs can name objects/actions but struggle to precisely detect "contact" and "release" events in videos, indicating a lack of perceptual grounding.
HCI predict mental health by encoding dimensions of self-reported states and their changes, enabling passive, scalable assessments.




HCI predict mental health by encoding dimensions of self-reported states and their changes, enabling passive, scalable assessments.
Optimize pathogen signal: SRP54 binding & modeling enhance antigen expression for vaccine design.




Optimize pathogen signal: SRP54 binding & modeling enhance antigen expression for vaccine design.
High-throughput Candida/C. elegans pathogenicity via ML viability scoring.


High-throughput Candida/C. elegans pathogenicity via ML viability scoring.
Akkermansia phylogroups link to PD-1 blockade response in NSCLC, esp. AmIa.



Akkermansia phylogroups link to PD-1 blockade response in NSCLC, esp. AmIa.
AD: Immune shift varies w/ age; tolerance(kids) to inflamm(adults) & innate sig(elderly).




AD: Immune shift varies w/ age; tolerance(kids) to inflamm(adults) & innate sig(elderly).
Models ultra-long eccDNA via bidirectional SSM & circ. augment. to boost cancer tasks.




Models ultra-long eccDNA via bidirectional SSM & circ. augment. to boost cancer tasks.
Single-cell RNA-seq analysis ID's cell-spec. genes/pathways in Alzheimers.




Single-cell RNA-seq analysis ID's cell-spec. genes/pathways in Alzheimers.
JADE aligns spatial transcriptomics slices using attention, iteratively refining alignment and embeddings for robust integration.




JADE aligns spatial transcriptomics slices using attention, iteratively refining alignment and embeddings for robust integration.
Region-based Deep Learn: B-cell Epitope Prediction.
Uses prot. lang. models + equivariant graph nets to cluster spatial epitope residues.




Region-based Deep Learn: B-cell Epitope Prediction.
Uses prot. lang. models + equivariant graph nets to cluster spatial epitope residues.
Genotype-phenotype prediction via hierarchical graph neural nets models gene interactions w/ attention.




Genotype-phenotype prediction via hierarchical graph neural nets models gene interactions w/ attention.




Maps condensates w/ microfluidics & lysate. Probes stability in stress granules, nucleoli, & FUS mutants.


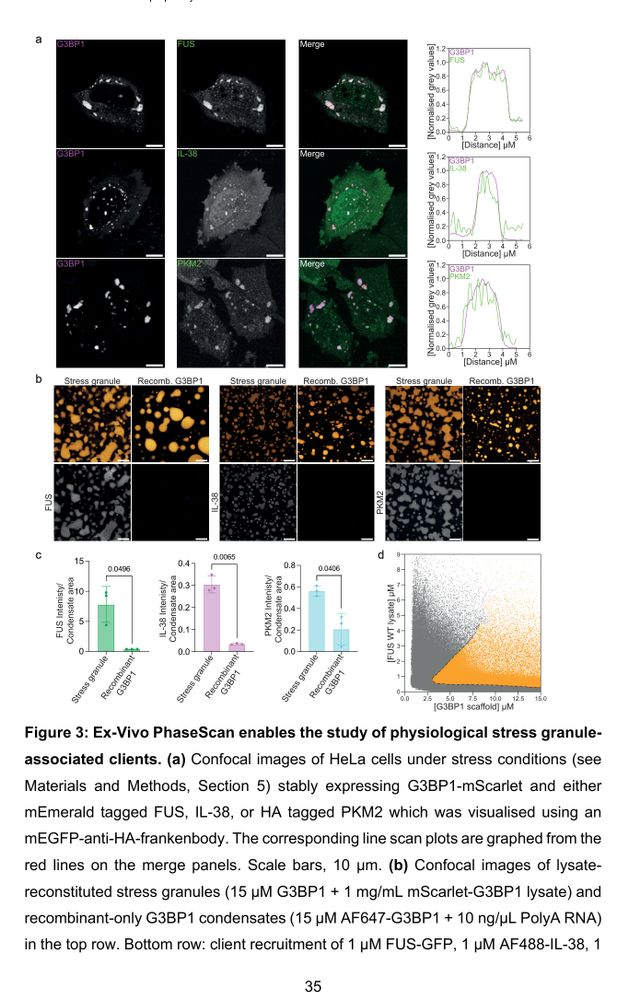

Maps condensates w/ microfluidics & lysate. Probes stability in stress granules, nucleoli, & FUS mutants.
*De novo* protein design potential via LLMs; promising for engineering.



*De novo* protein design potential via LLMs; promising for engineering.
Agentic LLM framework aids mechanistic enzyme design by integrating reasoning, structural analysis, and property prediction to generate testable hypotheses.




Agentic LLM framework aids mechanistic enzyme design by integrating reasoning, structural analysis, and property prediction to generate testable hypotheses.
Generates protein backbones by denoising torsion angles, ensuring ideal local geometry. Refinement optimizes compactness while maintaining bond constraints.




Generates protein backbones by denoising torsion angles, ensuring ideal local geometry. Refinement optimizes compactness while maintaining bond constraints.
GFlowNets open box: learns chem rules, finds key atomic env, prop, motifs in mol design.




GFlowNets open box: learns chem rules, finds key atomic env, prop, motifs in mol design.
Infers cell lineage via SNVs & CNAs jointly for accuracy.




Infers cell lineage via SNVs & CNAs jointly for accuracy.
Inferring cell environments by modeling cell interactions as agents learned via reinforcement learning. Enables in-silico manipulation and prediction.




Inferring cell environments by modeling cell interactions as agents learned via reinforcement learning. Enables in-silico manipulation and prediction.

