Koen De Gelas
@kdegelas.bsky.social
150 followers
500 following
36 posts
STA @ 10X Genomics, views my own
Posts
Media
Videos
Starter Packs
Reposted by Koen De Gelas
Reposted by Koen De Gelas
Reposted by Koen De Gelas
Koen De Gelas
@kdegelas.bsky.social
· Aug 14

Plant-Compatible Xenium In Situ Sequencing: Optimised Protocol for Spatial Transcriptomics in Medicago truncatula Roots and Nodules
Elucidating the spatial and temporal regulation of gene expression during plant organogenesis is crucial for enabling precise crop improvement strategies that incorporate beneficial traits into crops ...
www.biorxiv.org
Koen De Gelas
@kdegelas.bsky.social
· Aug 7
Koen De Gelas
@kdegelas.bsky.social
· Aug 7
Koen De Gelas
@kdegelas.bsky.social
· Jul 29

One Section, Two Worlds: Single-Cell Integration of MALDI-MSI and Spatial Transcriptomics on the Same Single Tissue Section
Understanding tissue complexity requires spatially resolved multiomic data at single-cell resolution. Here, we present a workflow that integrates high-resolution matrix-assisted laser desorption ioniz...
www.biorxiv.org
Reposted by Koen De Gelas
Koen De Gelas
@kdegelas.bsky.social
· Jun 23
Karsten Rippe
@karsten-rippe.bsky.social
· Jun 20
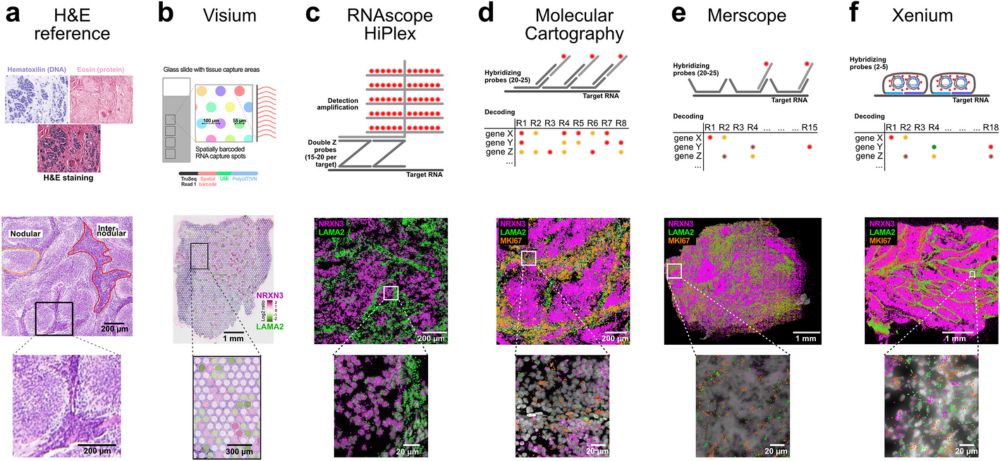
Comparison of spatial transcriptomics technologies using tumor cryosections - Genome Biology
Background Spatial transcriptomics technologies are revolutionizing our understanding of intra-tumor heterogeneity and the tumor microenvironment by revealing single-cell molecular profiles within the...
doi.org
Reposted by Koen De Gelas
Reposted by Koen De Gelas
Reposted by Koen De Gelas
Koen De Gelas
@kdegelas.bsky.social
· Jun 6

How mapping the spatial biology of the tumor microenvironment can benefit cancer drug discovery and development - 10x Genomics
Spatial transcriptomics reveals complex and therapeutically relevant interactions among tumor and immune cells in the TME and tumor boundary.
bit.ly
Koen De Gelas
@kdegelas.bsky.social
· Jun 6

High-definition spatial transcriptomic profiling of immune cell populations in colorectal cancer - Nature Genetics
High-resolution, spatial transcriptomic analysis of colorectal cancers using matched sequencing- and imaging-based methods characterizes immune cells and their interactions with the tumor microenviron...
www.nature.com
Reposted by Koen De Gelas
Reposted by Koen De Gelas