
Website: https://steglelab.org/
We’re excited to welcome Prof. Dr. Oliver Stegle as keynote speaker at #HMCConference2026
Director & Co-Spokesperson of #GHGA, @oliverstegle.bsky.social advances secure, #FAIR infrastructures for data-driven biomedical research. 🧬
steglelab.bsky.social ghga.bsky.social

We’re excited to welcome Prof. Dr. Oliver Stegle as keynote speaker at #HMCConference2026
Director & Co-Spokesperson of #GHGA, @oliverstegle.bsky.social advances secure, #FAIR infrastructures for data-driven biomedical research. 🧬
steglelab.bsky.social ghga.bsky.social
Preprint: doi.org/10.64898/202...
Summary in the 🧵 below by @danaivagiaki.bsky.social
Preprint: doi.org/10.64898/202...
Summary in the 🧵 below by @danaivagiaki.bsky.social
🔗 Deadline: 15/02/2026
embl.wd103.myworkdayjobs.com/en-US/EMBL/d...

🔗 Deadline: 15/02/2026
embl.wd103.myworkdayjobs.com/en-US/EMBL/d...
For more information, see the @hlsalliance.bsky.social Postdoc Program:
www.health-life-sciences.de/opportunitie...
For more information, see the @hlsalliance.bsky.social Postdoc Program:
www.health-life-sciences.de/opportunitie...
Register now and submit your poster: elsa-ai.eu/elsa-worksho...
📅 March 9, Heidelberg DE
Register now and submit your poster: elsa-ai.eu/elsa-worksho...
📅 March 9, Heidelberg DE
Take a look at our recent perspective: "Interpretation, extrapolation and perturbation of single cells"! (rdcu.be/eXeDY)

Take a look at our recent perspective: "Interpretation, extrapolation and perturbation of single cells"! (rdcu.be/eXeDY)
Most single-cell CRISPR screens infer edits indirectly.
We present scPT-seq, a method that directly detects CRISPR mutations and transcriptomes in the same single cells in vivo, from John Hawkins and Siamak Redhai from Boutros Lab. 1/10

Most single-cell CRISPR screens infer edits indirectly.
We present scPT-seq, a method that directly detects CRISPR mutations and transcriptomes in the same single cells in vivo, from John Hawkins and Siamak Redhai from Boutros Lab. 1/10
🔗 Apply here: jobs.dkfz.de/en/jobs/1683...

🔗 Apply here: jobs.dkfz.de/en/jobs/1683...
📅 March 9, 2026
📍 EMBL, Heidelberg, Germany
Join us on March 9, 2026, to advance private, fair, robust, reliable, and accountable AI models for healthcare applications.
elsa-ai.eu/elsa-worksho...

📅 March 9, 2026
📍 EMBL, Heidelberg, Germany
Hendrik’s research focuses on reliable machine learning methods integrating large-scale EHR data with multimodal data like genomics data, with a focus on causality and uncertainty estimation.

Hendrik’s research focuses on reliable machine learning methods integrating large-scale EHR data with multimodal data like genomics data, with a focus on causality and uncertainty estimation.

We also presented our latest findings from the lab 👇

We also presented our latest findings from the lab 👇
Read the full article: ellis.eu/news/ellis-s...

Read the full article: ellis.eu/news/ellis-s...
During her molecular biology studies, Gesa developed an interest in applying mathematics and ML to understand complex biological systems. Her PhD research will focus on inferring GRNs by integrating insights from population genetics and genetic perturbation data.

During her molecular biology studies, Gesa developed an interest in applying mathematics and ML to understand complex biological systems. Her PhD research will focus on inferring GRNs by integrating insights from population genetics and genetic perturbation data.
Steffi joins us as a PhD student after completing her Master’s at Heidelberg University. She will explore gene regulation and cell differentiation using organoid models and is excited to analyze these complex datasets and develop ML methods.

Steffi joins us as a PhD student after completing her Master’s at Heidelberg University. She will explore gene regulation and cell differentiation using organoid models and is excited to analyze these complex datasets and develop ML methods.
You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n

You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n
Led by EMBL’s Oliver Stegle and Anna Kreshuk, the workshop united researchers and AI experts to explore how advanced computational approaches are shaping the future of life sciences.
#ai

Led by EMBL’s Oliver Stegle and Anna Kreshuk, the workshop united researchers and AI experts to explore how advanced computational approaches are shaping the future of life sciences.
#ai
We’ll generate high-throughput multi-modal CRISPR data on ASD risk genes in 🧠 organoids & assembloids, and build an ML model of gene regulation dynamics to analyze the data and enable future in silico experiments.
👉TACTICS: Thalamocortical Assembloids and Computational Tools for Investigating Cellular Signatures in #Autism.
🧠How do autism-linked mutations disrupt early brain development? @moritzmall.bsky.social @steglelab.bsky.social
@dkfz.bsky.social

We’ll generate high-throughput multi-modal CRISPR data on ASD risk genes in 🧠 organoids & assembloids, and build an ML model of gene regulation dynamics to analyze the data and enable future in silico experiments.
SDR-seq makes it possible; an experimental method connecting coding and non-coding variants to expression in single cells.
Proud to have contributed a custom FASTQ-processing tool to this great collaborative project.
Known as SDR-seq, the tool has unprecedented scale, precision, and speed to characterise variants and link them to functions and disease.
www.embl.org/news/science...

SDR-seq makes it possible; an experimental method connecting coding and non-coding variants to expression in single cells.
Proud to have contributed a custom FASTQ-processing tool to this great collaborative project.
📅 October 16, 2:30pm-4:30pm
📍 Poster 1007T
📅 October 16, 2:30pm-4:30pm
📍 Poster 1007T
A fantastic opportunity to showcase and discover innovative data frameworks for #singlecell analysis, and to meet the developers behind the scverse ecosystem. Looking forward to connecting with the community!
Register now! 👇
A fantastic opportunity to showcase and discover innovative data frameworks for #singlecell analysis, and to meet the developers behind the scverse ecosystem. Looking forward to connecting with the community!
Register now! 👇
Check out the publication for the nuts and bolts of 🍫-G2P: doi.org/10.1038/s415...
Clever experimental design by @breinigmarco.bsky.social hijacking Visium and elegant analysis by @lomakinai.bsky.social and @elihei.bsky.social
www.nature.com/articles/s41...
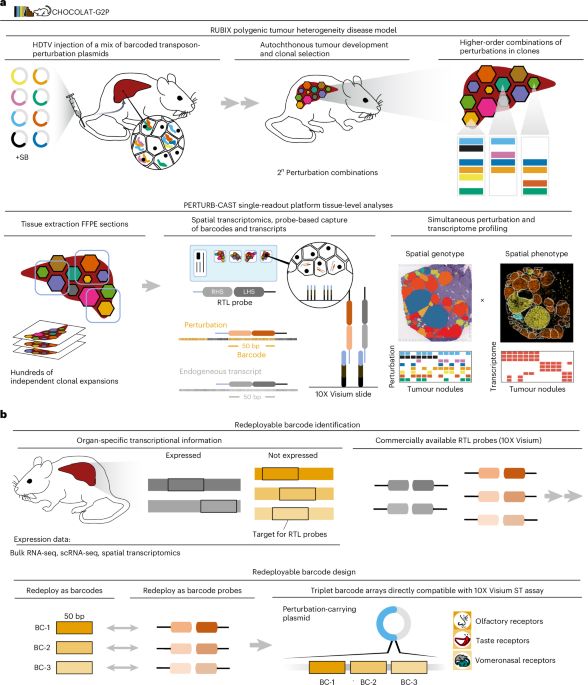
Check out the publication for the nuts and bolts of 🍫-G2P: doi.org/10.1038/s415...
You'll be working on multi-omics factorization models for personalized medicine, co-supervised by @junyanlu.bsky.social.
👉 Project: www.helmholtz.de/assets/hidss...
✍️ Apply: hidss4health.de/application

You'll be working on multi-omics factorization models for personalized medicine, co-supervised by @junyanlu.bsky.social.
👉 Project: www.helmholtz.de/assets/hidss...
✍️ Apply: hidss4health.de/application
💡 Funding from the Hector Foundation is dedicated to attracting interdisciplinary talent, boosting infrastructure, training, & innovation.
www.embl.org/news/connect...

💡 Funding from the Hector Foundation is dedicated to attracting interdisciplinary talent, boosting infrastructure, training, & innovation.
www.embl.org/news/connect...
has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...

has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...

