Timothy P. Jenkins
@tpjenkins.bsky.social
470 followers
1.3K following
77 posts
I am an Associate Professor at DTU and leading the Digital Biotechnology Lab. I am passionate about harnessing the power of modern technology such as AI Protein Design to innovate the biotechnological landscape and develop real world solutions.
Posts
Media
Videos
Starter Packs
Timothy P. Jenkins
@tpjenkins.bsky.social
· Jul 16
Timothy P. Jenkins
@tpjenkins.bsky.social
· Jun 11
Reposted by Timothy P. Jenkins
Timothy P. Jenkins
@tpjenkins.bsky.social
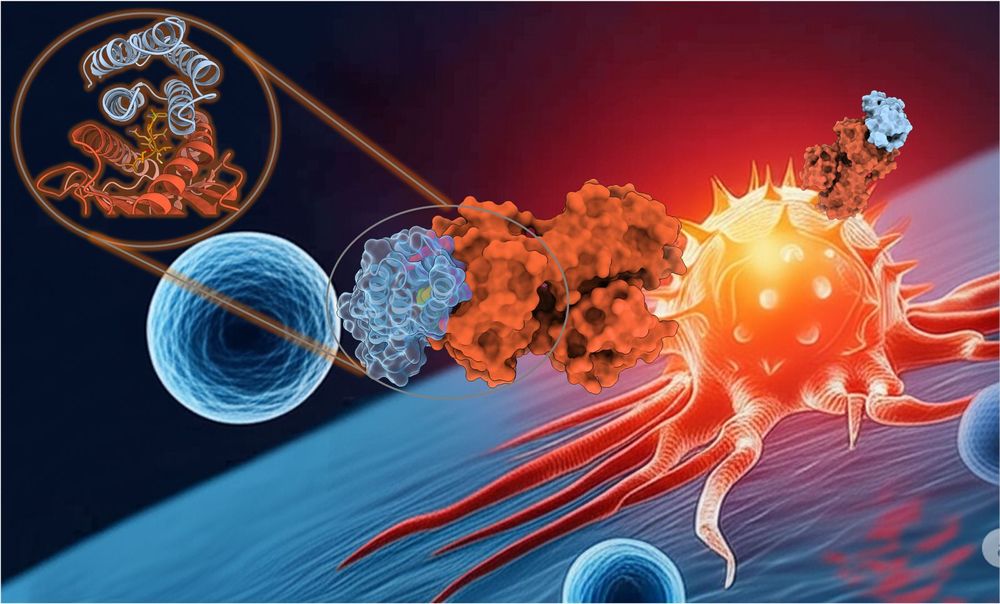
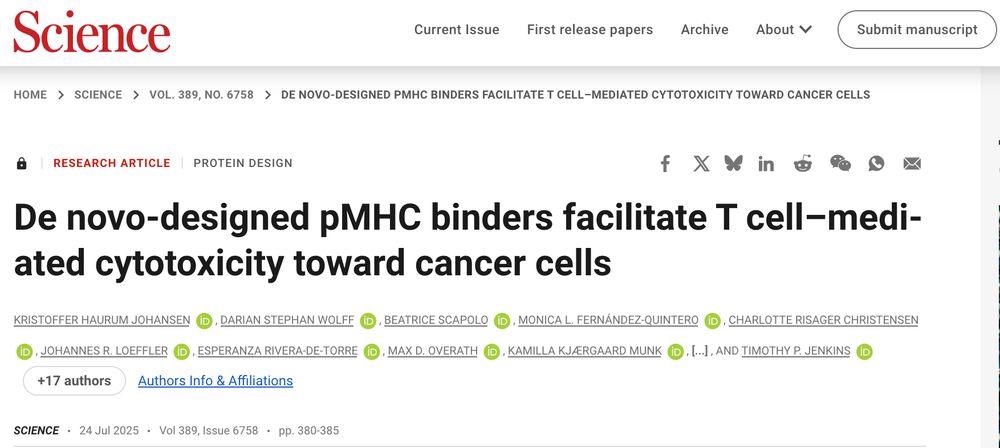
· May 30
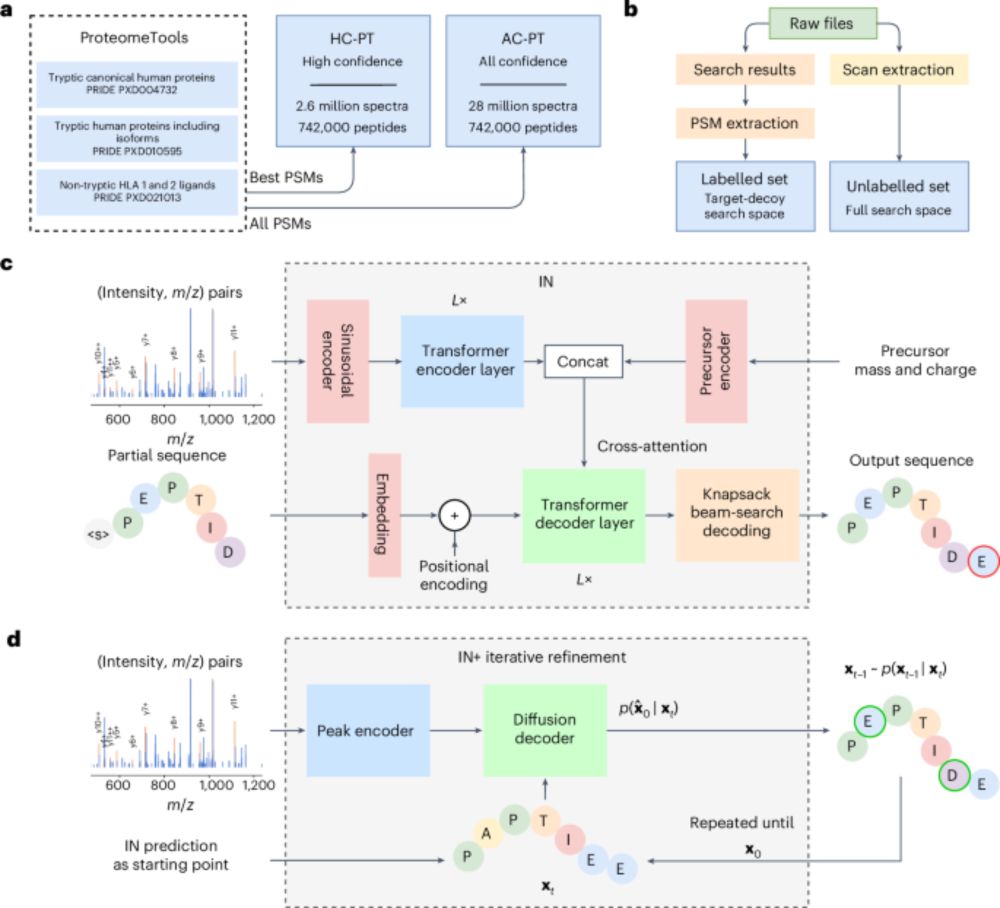
InstaNovo enables diffusion-powered de novo peptide sequencing in large-scale proteomics experiments - Nature Machine Intelligence
InstaNovo, a transformer-based model, and InstaNovo+, a multinomial diffusion model, enhance de novo peptide sequencing, enabling discovery of novel peptides, improved therapeutics sequencing coverage...
www.nature.com
Reposted by Timothy P. Jenkins
Reposted by Timothy P. Jenkins