Dominik Handler
@86dominik.bsky.social
260 followers
360 following
19 posts
Drosophila genetic conflicts
piRNAs | transposons | genomics
Staff scientist in the Brennecke lab - Vienna
Posts
Media
Videos
Starter Packs
Reposted by Dominik Handler
Thrilled to share that I’ll be joining @imbmainz.bsky.social in February 2026 to start my own group!
We will explore new mechanisms in eukaryotic gene expression, leveraging ‘evolutionary play’ to uncover how regulation, repurposing, and hijacking shape RNA biology.
PhD positions available!
We will explore new mechanisms in eukaryotic gene expression, leveraging ‘evolutionary play’ to uncover how regulation, repurposing, and hijacking shape RNA biology.
PhD positions available!
Reposted by Dominik Handler
Dominik Handler
@86dominik.bsky.social
· Aug 21
Reposted by Dominik Handler
Reposted by Dominik Handler
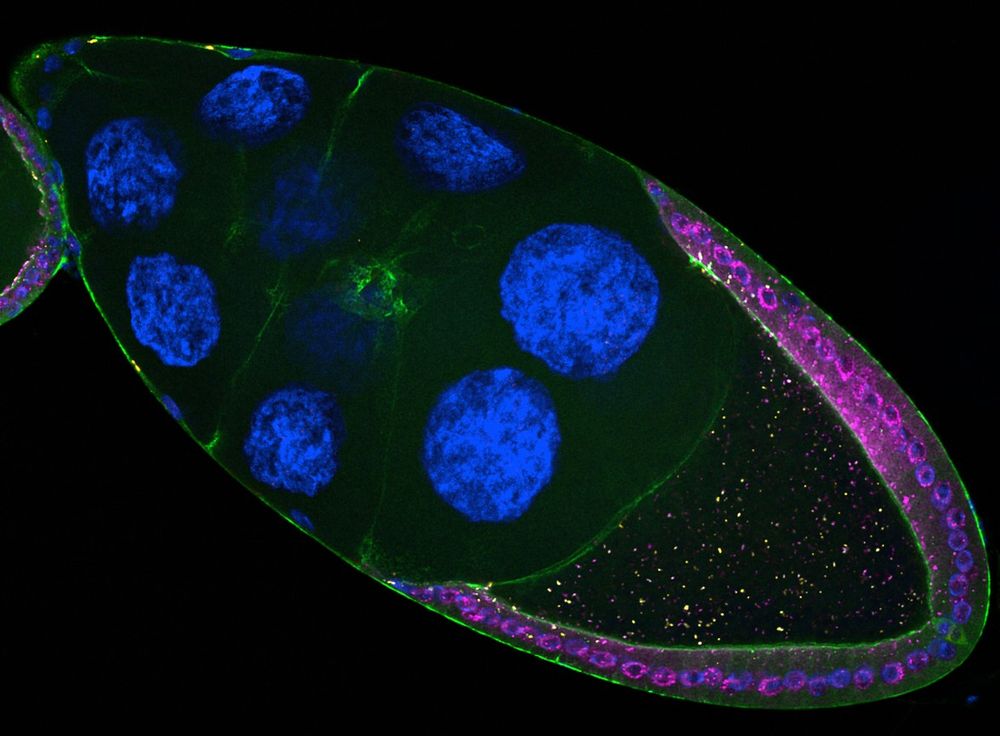
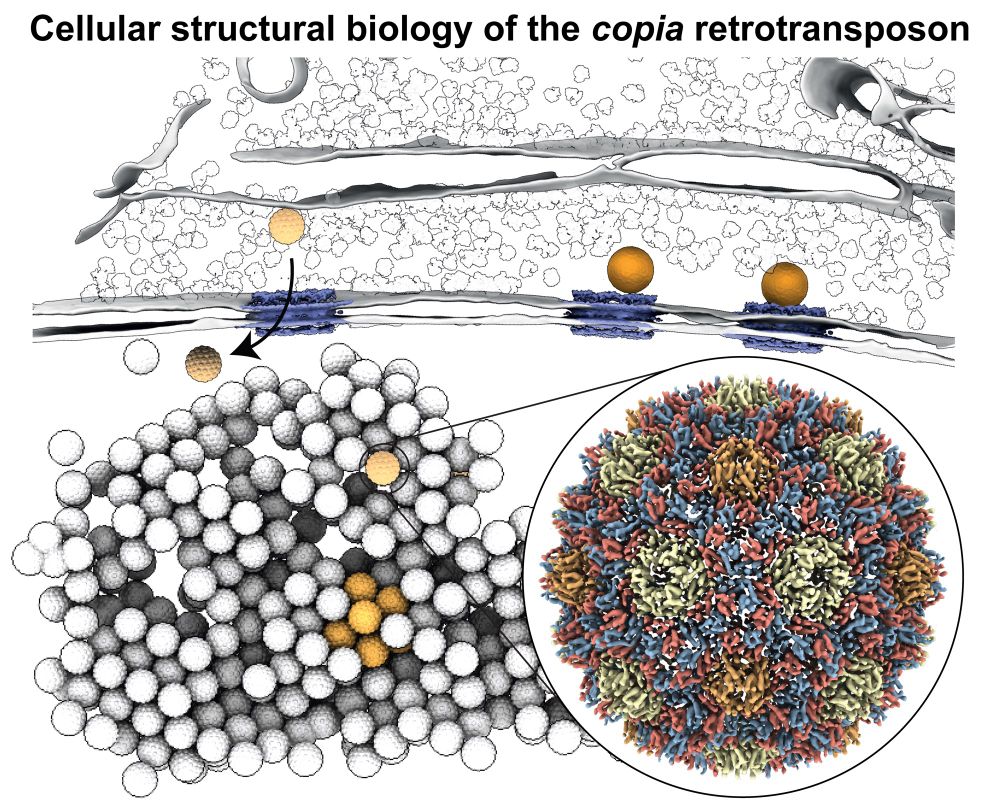
Antisense transposon insertions into host genes trigger piRNA mediated immunity https://www.biorxiv.org/content/10.1101/2025.07.28.667215v1
Reposted by Dominik Handler
Reposted by Dominik Handler
IMBA is recruiting a Junior Group Leader! Are you interested in starting your own lab, pursuing curiosity-driven basic research in the life sciences? Apply now to our group leader position. The deadline is May 28. Link is below.

Reposted by Dominik Handler
Reposted by Dominik Handler