
🔗 doi.org/10.1093/molbev/msaf257
#evobio #molbio #compbio #popgen

🔗 doi.org/10.1093/molbev/msaf257
#evobio #molbio #compbio #popgen
@tylervkent.bsky.social @samurscicop.bsky.social & D. Matute analysed 131 genomes of the globally invasive Aedes aegypti, and found high genomic resilience despite historical eradication attempts for this globally invasive vector.
🔗 doi.org/10.1093/gbe/...
#genome #mosquitoes
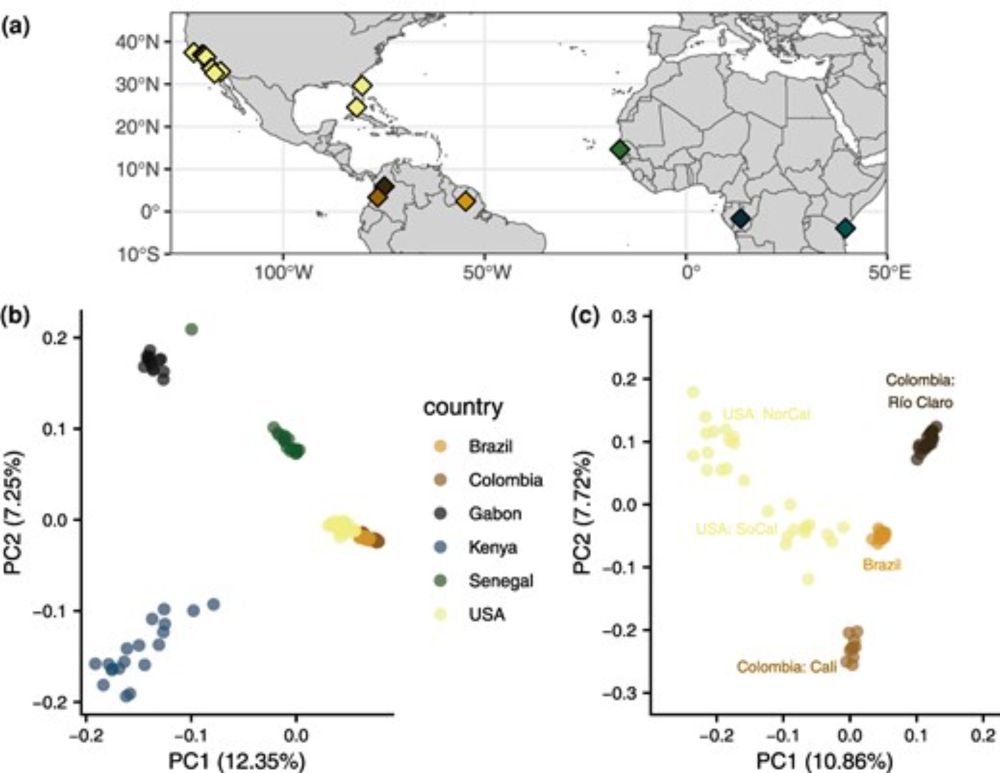
@tylervkent.bsky.social @samurscicop.bsky.social & D. Matute analysed 131 genomes of the globally invasive Aedes aegypti, and found high genomic resilience despite historical eradication attempts for this globally invasive vector.
🔗 doi.org/10.1093/gbe/...
#genome #mosquitoes

We took a dive into the complicated demographic history of Aedes aegypti mosquitoes and its impact on the distribution of genetic diversity
doi.org/10.1093/gbe/...

We took a dive into the complicated demographic history of Aedes aegypti mosquitoes and its impact on the distribution of genetic diversity
doi.org/10.1093/gbe/...


doi.org/10.1093/evol...
@journal-evo.bsky.social #popgen #evobio

doi.org/10.1093/evol...
@journal-evo.bsky.social #popgen #evobio


