

github.com/acnash
"Data-Centric Benchmarking of Neural Network Architectures for the Univariate Time Series Forecasting Task"
#timeseries #LSTM #realworlddata #neuralnetworks
"Data-Centric Benchmarking of Neural Network Architectures for the Univariate Time Series Forecasting Task"
#timeseries #LSTM #realworlddata #neuralnetworks
It includes comprehensive MD tutorials in GROMACS, covering forcefields, thermodynamic ensembles, long-range electrostatics and much more!

It includes comprehensive MD tutorials in GROMACS, covering forcefields, thermodynamic ensembles, long-range electrostatics and much more!

Upgrading GROMACS to handle billion-atom systems and enhancing I/O performance and precision, making the first-ever whole-cell simulation possible ➡️ bioexcel.eu/uw67
#MolecularDynamics #GROMACS #ComputationalBiology

Upgrading GROMACS to handle billion-atom systems and enhancing I/O performance and precision, making the first-ever whole-cell simulation possible ➡️ bioexcel.eu/uw67
#MolecularDynamics #GROMACS #ComputationalBiology
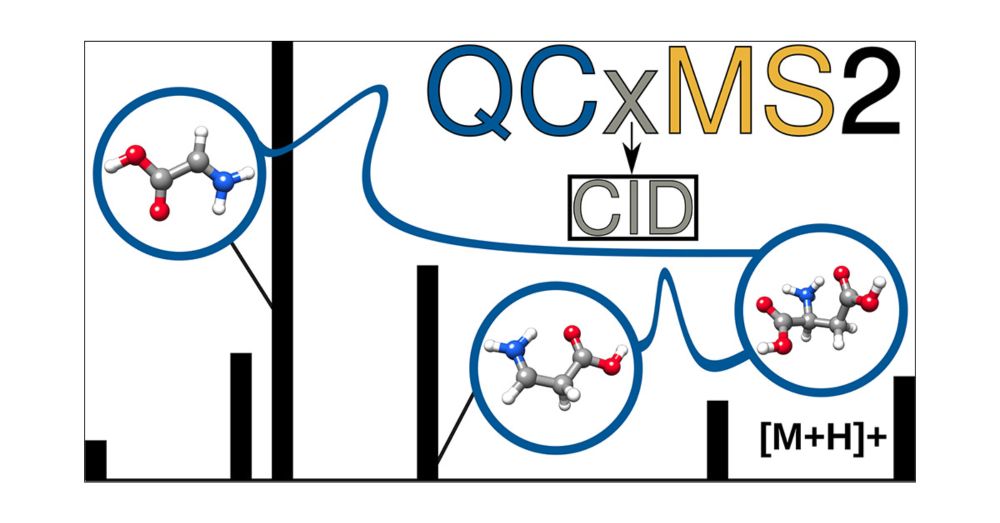
Just published in #JASMS : doi.org/10.1021/jasms.5c00234
Grateful to my coauthors Stefan Grimme @grimmelab.bsky.social & Marianne Engeser @unibonn.bsky.social - this is the last project of my PhD and completes my work on QCxMS2!
#MassSpec #compchem
Just published in #JASMS : doi.org/10.1021/jasms.5c00234
Grateful to my coauthors Stefan Grimme @grimmelab.bsky.social & Marianne Engeser @unibonn.bsky.social - this is the last project of my PhD and completes my work on QCxMS2!
#MassSpec #compchem


From now on, I'm painting, playing games, and travelling 😀
From now on, I'm painting, playing games, and travelling 😀
github.com/acnash

github.com/acnash


This is going to break. I just know it.
#sciencesoftware
This is going to break. I just know it.
#sciencesoftware
Or why automated virtual screening and docking remains hard and why expertise remains essential. medchemash.substack.com/p/on-the-fai...

Or why automated virtual screening and docking remains hard and why expertise remains essential. medchemash.substack.com/p/on-the-fai...
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
doi.org/10.1093/bioi...
VTX is open source and freely accessible for non commercial use! github.com/VTX-Molecula...
Builds available at vtx.drugdesign.fr
Organisational skills must be at another level.
huggingface.co/facebook/OMo...
And the paper:
arxiv.org/abs/2505.08762

Organisational skills must be at another level.
huggingface.co/facebook/OMo...
And the paper:
arxiv.org/abs/2505.08762
Some tips for performing meaningful and reproducible docking calculations.
journals.plos.org/ploscompbiol...
#docking #moleculardocking #liganddocking #compchem

Some tips for performing meaningful and reproducible docking calculations.
journals.plos.org/ploscompbiol...
#docking #moleculardocking #liganddocking #compchem
#AI #ML #machinelearning #science
www.nature.com/articles/s41...

#AI #ML #machinelearning #science
www.nature.com/articles/s41...
Danum Anthony
www.dahliaworld.co.uk/dnamesv.htm#A
Danum Anthony
www.dahliaworld.co.uk/dnamesv.htm#A




