
• High-throughput approach to generate genome-wide loss-of-function phage libraries.
• Interrogates gene essentiality w/insights into phage genome organization, host antiviral defense & gene function
www.cell.com/cell-host-mi...
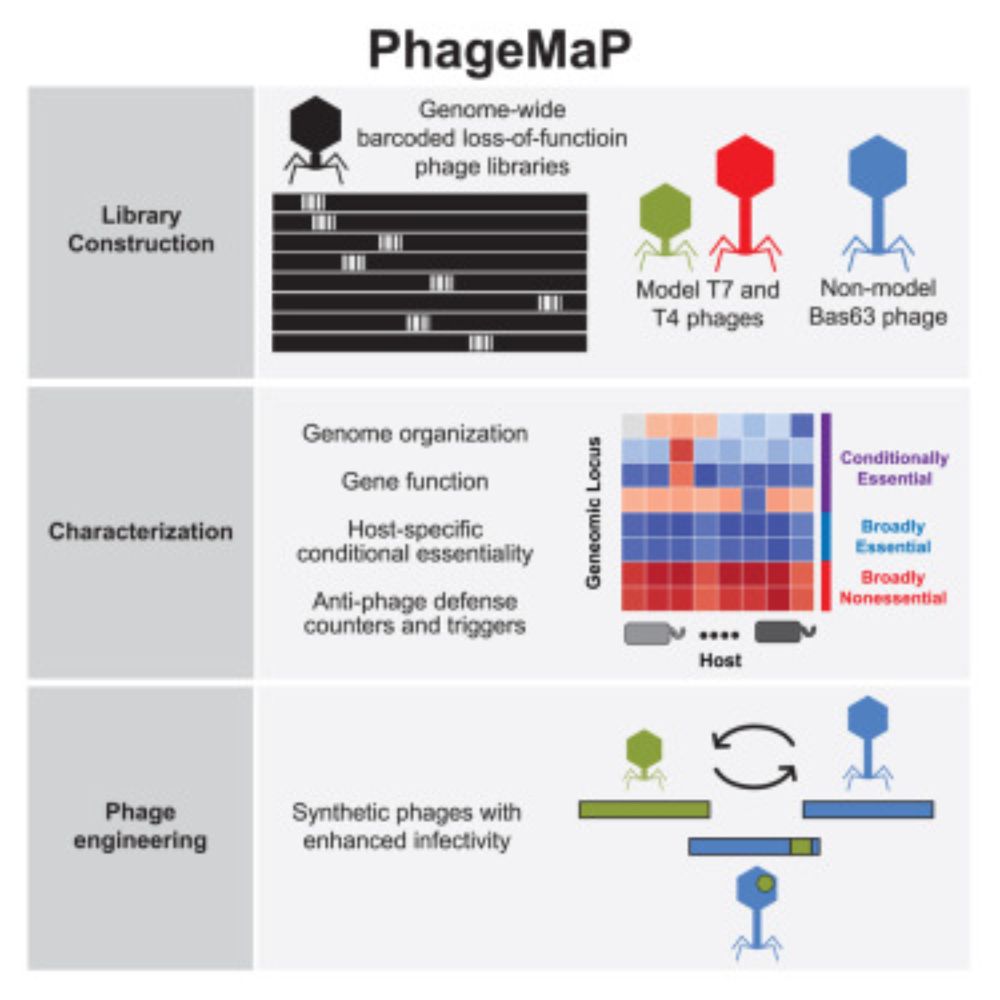
• High-throughput approach to generate genome-wide loss-of-function phage libraries.
• Interrogates gene essentiality w/insights into phage genome organization, host antiviral defense & gene function
www.cell.com/cell-host-mi...
journals.asm.org/doi/10.1128/... 🧵👇

journals.asm.org/doi/10.1128/... 🧵👇
We had a huge task—picking bacterial stocks from >200 96-well plates and re-arranging them—and the lab's anaerobic liquid handler was broken without the funds to repair. 🧵

We had a huge task—picking bacterial stocks from >200 96-well plates and re-arranging them—and the lab's anaerobic liquid handler was broken without the funds to repair. 🧵

A new publication, describing genome-scale resources in bifidobacteria, is now available online: doi.org/10.1016/j.ce...
#microbiome #microsky

A new publication, describing genome-scale resources in bifidobacteria, is now available online: doi.org/10.1016/j.ce...
#microbiome #microsky

