http://chaohou.netlify.app
#ProteinLM

#ProteinLM
doi.org/10.1101/2025...

doi.org/10.1101/2025...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
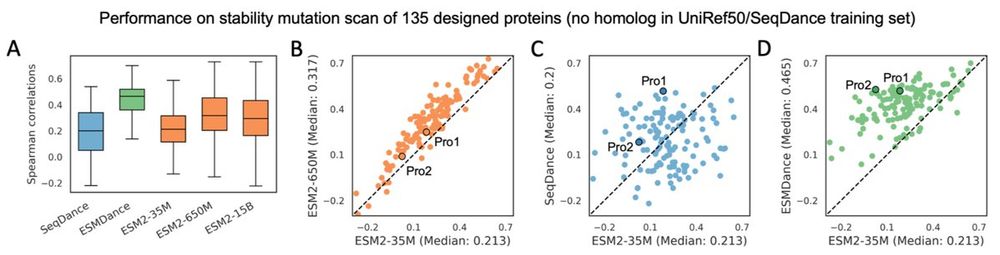
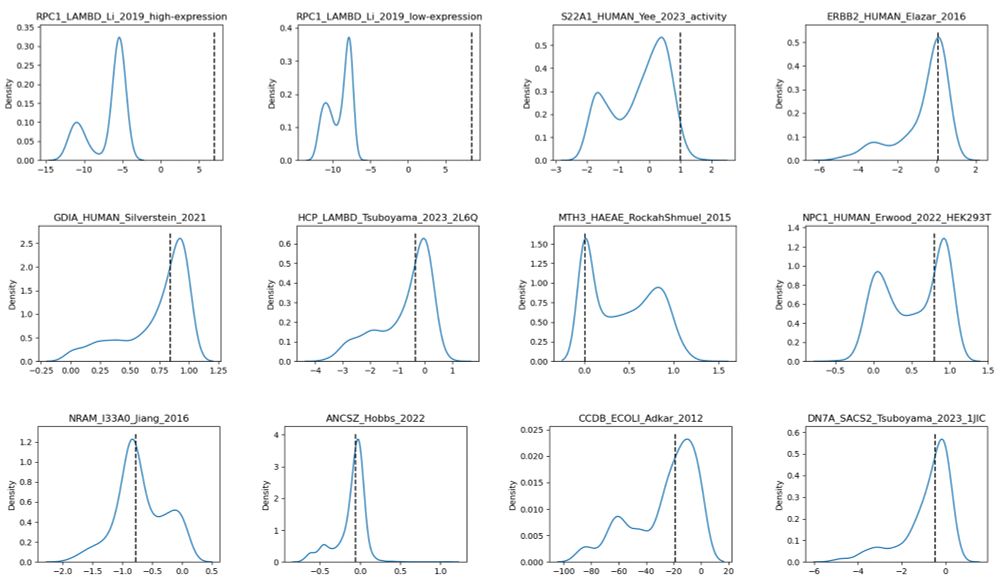
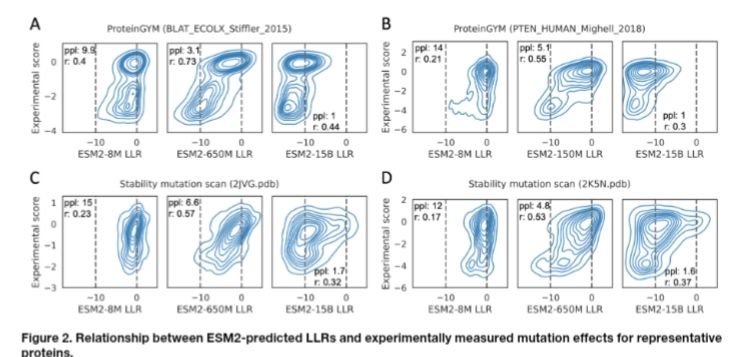
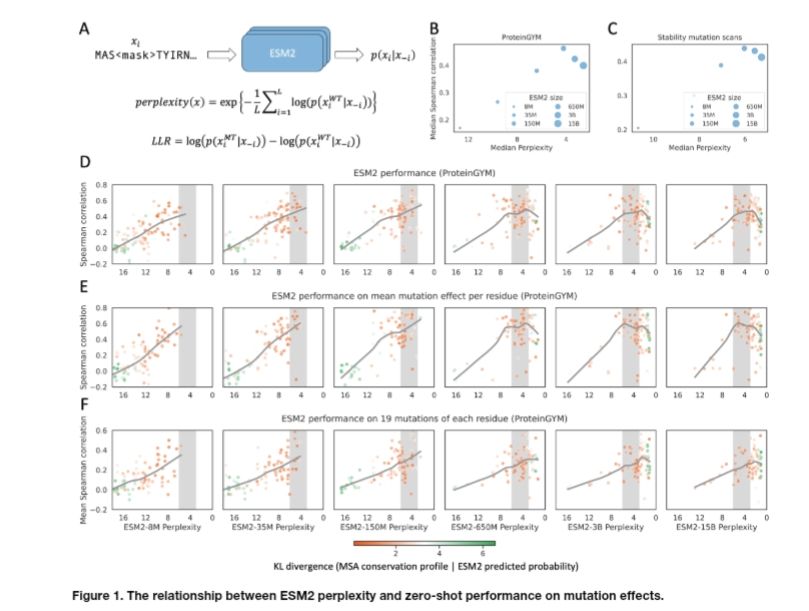
We dive into this issue in our new preprint—bringing insights into model scaling on mutation effect prediction. 🧬📉
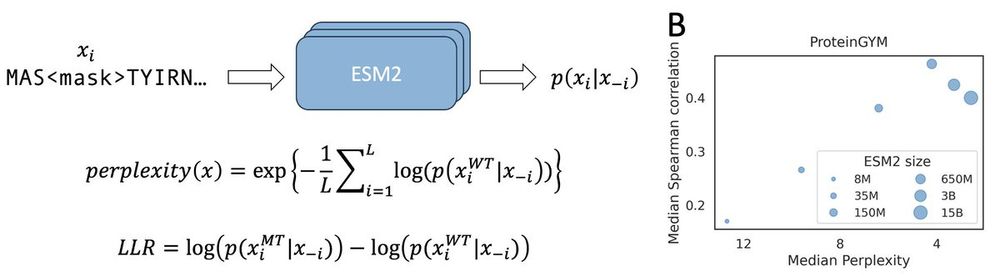
We dive into this issue in our new preprint—bringing insights into model scaling on mutation effect prediction. 🧬📉