
From France, raised in Madagascar and now in Edinburgh!
A long overdue study on what reference panel to use for optimal genotype imputation and genomic prediction from the #AquaImpact project
In silico imputation from 500 SNPs to 45K SNPs in Atlantic salmon was tested using many scenarios
🧬 💻
🐟
🧪👩🔬
www.biorxiv.org/content/10.1...

A long overdue study on what reference panel to use for optimal genotype imputation and genomic prediction from the #AquaImpact project
In silico imputation from 500 SNPs to 45K SNPs in Atlantic salmon was tested using many scenarios
🧬 💻
🐟
🧪👩🔬
www.biorxiv.org/content/10.1...
Delighted to see this manuscript on AGD in salmon Investigated Through a Holo‐Omic Lens from Eiríkur Thormar's time in Roslin a few years ago published in Ecology and Evolution
onlinelibrary.wiley.com/doi/10.1002/...
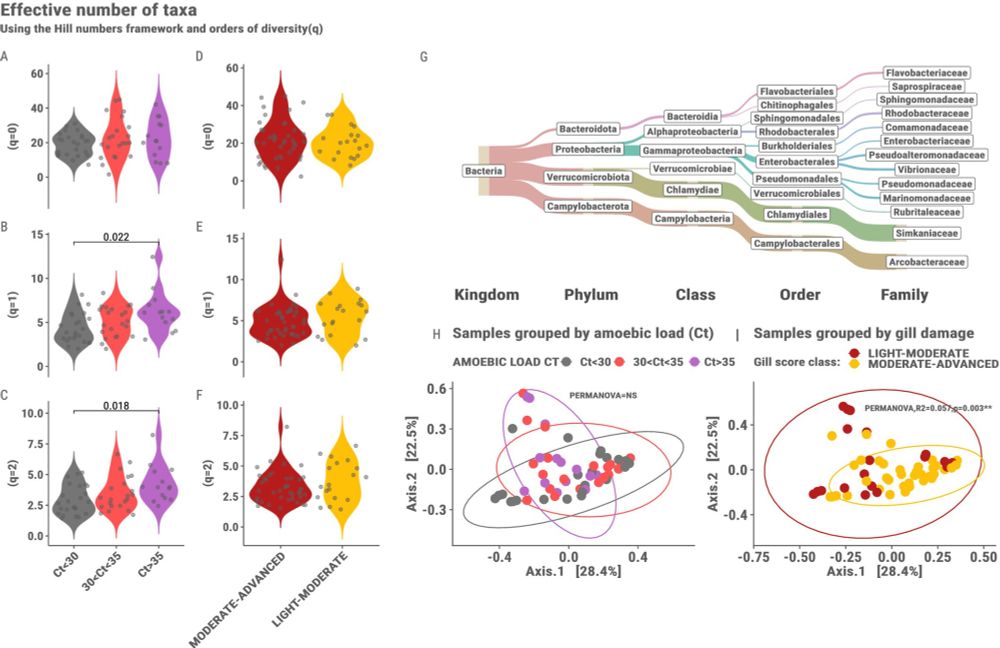
Delighted to see this manuscript on AGD in salmon Investigated Through a Holo‐Omic Lens from Eiríkur Thormar's time in Roslin a few years ago published in Ecology and Evolution
onlinelibrary.wiley.com/doi/10.1002/...
Well done Sarah !
#ISGAXV

Well done Sarah !
#ISGAXV

From SNP array development to implementation of genomic selection 🧬🖥️🐟
From SNP array development to implementation of genomic selection 🧬🖥️🐟
Today's plenary was an overview of oyster production and selection in France by Romain Morvezen
Today's plenary was an overview of oyster production and selection in France by Romain Morvezen
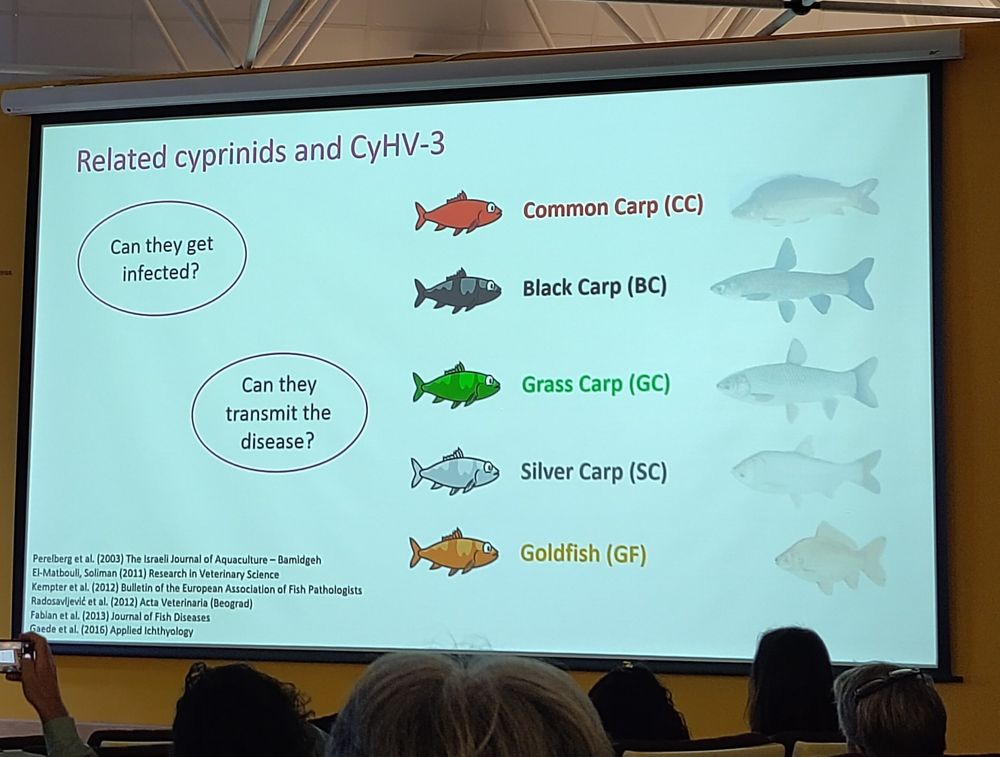
We heard from Duwage C.G. Rodrigo (Jeju Uni, Korea) about resistance to M avidus in olive flounders. Polygenic + heritable = genomic selection to improve resistance in the population ✅
We heard from Duwage C.G. Rodrigo (Jeju Uni, Korea) about resistance to M avidus in olive flounders. Polygenic + heritable = genomic selection to improve resistance in the population ✅
🧬🐟
🧬🐟
www.biorxiv.org/content/10.1...
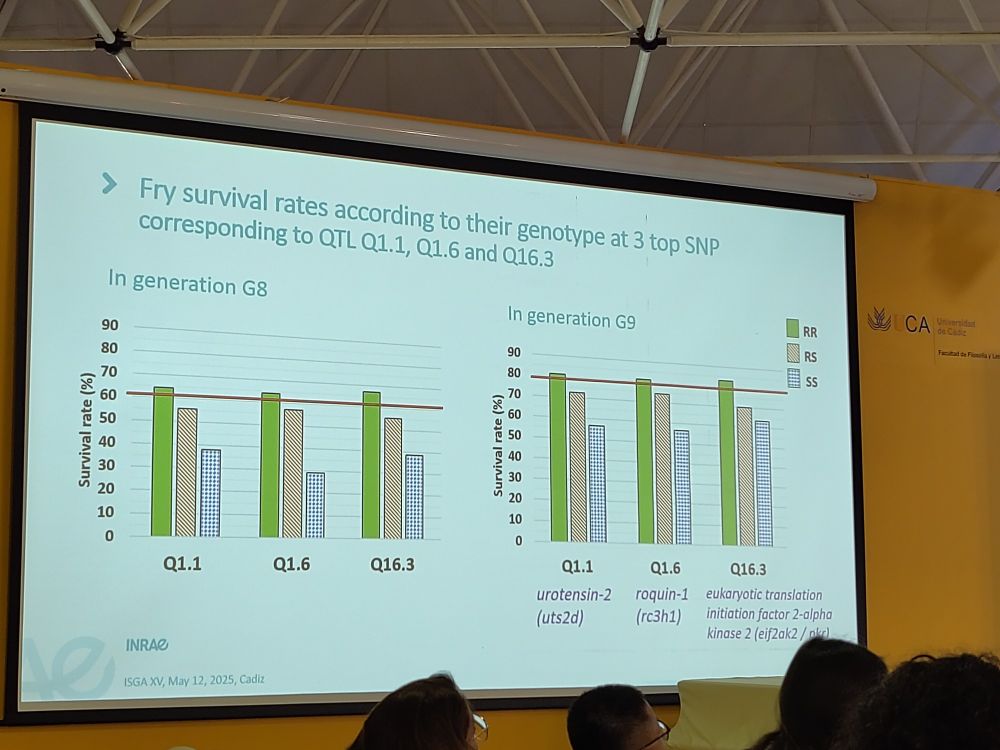
www.biorxiv.org/content/10.1...
Earlier there was a presentation by Pierrick Haffray (SYSAAF) on accounting for molt stages in genetic evaluation of 🔵🦐. Showing that time to molt was heritable but molt stages wasn't in their dataset (late measurement)
Earlier there was a presentation by Pierrick Haffray (SYSAAF) on accounting for molt stages in genetic evaluation of 🔵🦐. Showing that time to molt was heritable but molt stages wasn't in their dataset (late measurement)

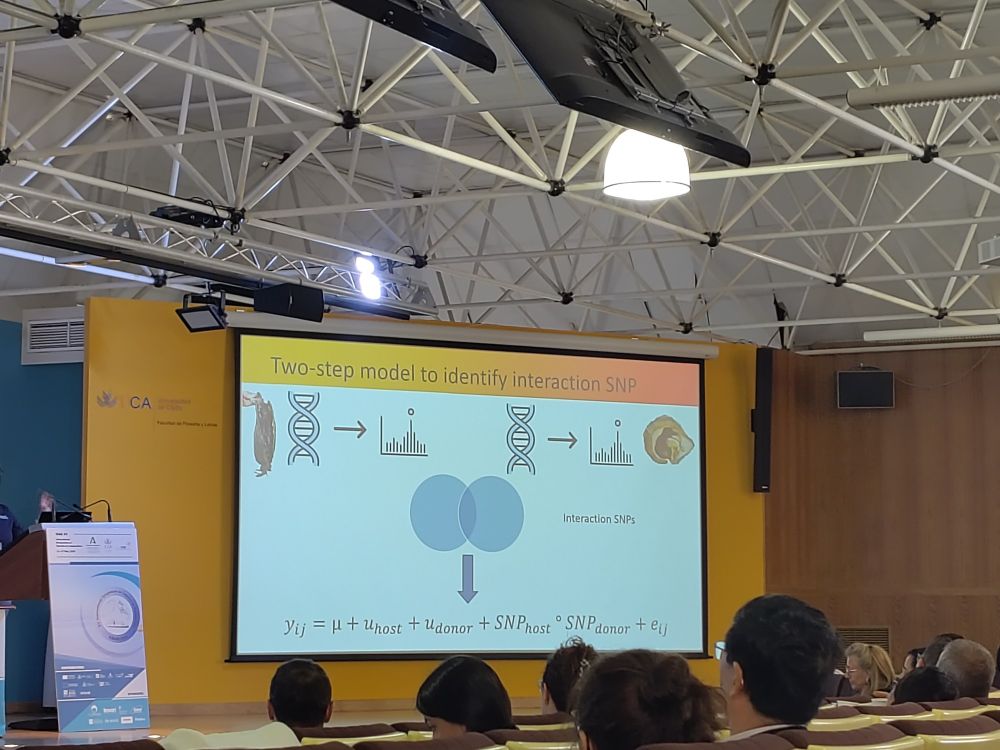
#ISGAXV
🧬🖥️ 🐟
#ISGAXV
🧬🖥️ 🐟
A frist study in this population from Canada.
A frist study in this population from Canada.


The conference started yesterday with a nice reception and it's great to see so many familiar faces and catch up with friends!
Today is the first day of the official business: talking about aquaculture, genetics and genomics
🧪
🐟
🖥️🧬
The conference started yesterday with a nice reception and it's great to see so many familiar faces and catch up with friends!
Today is the first day of the official business: talking about aquaculture, genetics and genomics
🧪
🐟
🖥️🧬


We finaly got our follow up study on fine mapping of QTL associated with spontaneous malness in XX-rainbow trout out in PlosOne!
Check it out: dx.plos.org/10.1371/jour...
It's a much improved version of the preprint (thanks to reviewers and editors and to Florence Phocas (INRAe)
We finaly got our follow up study on fine mapping of QTL associated with spontaneous malness in XX-rainbow trout out in PlosOne!
Check it out: dx.plos.org/10.1371/jour...
It's a much improved version of the preprint (thanks to reviewers and editors and to Florence Phocas (INRAe)
HOWEVER (and that is a big however) we are a long long way from gender balance... 🙄
👩🔬
HOWEVER (and that is a big however) we are a long long way from gender balance... 🙄
👩🔬
Using multi-omics approached they detected a major QTL and identified causative mutations in the promoter regions of IFI27L2A genes! 🧬💻 🐟 🧪

Using multi-omics approached they detected a major QTL and identified causative mutations in the promoter regions of IFI27L2A genes! 🧬💻 🐟 🧪
🧬🖥️

🧬🖥️

