William Beimers
@diomed.bsky.social
150 followers
160 following
14 posts
PhD Student in Biochemistry @ UW-Madison
focus on plasma proteomics technologies and applications
website: https://wbeimers.github.io/
Posts
Media
Videos
Starter Packs
Reposted by William Beimers
William Beimers
@diomed.bsky.social
· Sep 5
William Beimers
@diomed.bsky.social
· Sep 3

Impact of Sample Preparation Strategies on the Quantitative Accuracy of Low-Abundance Serum Proteins in Shotgun Proteomics
Serum proteomics plays a crucial role in biomarker discovery and disease research, yet the selection of an optimal sample preparation method remains challenging. Evaluating the accuracy of protein qua...
pubs.acs.org
William Beimers
@diomed.bsky.social
· Jul 18
Reposted by William Beimers
William Beimers
@diomed.bsky.social
· May 14
William Beimers
@diomed.bsky.social
· May 14
William Beimers
@diomed.bsky.social
· May 14
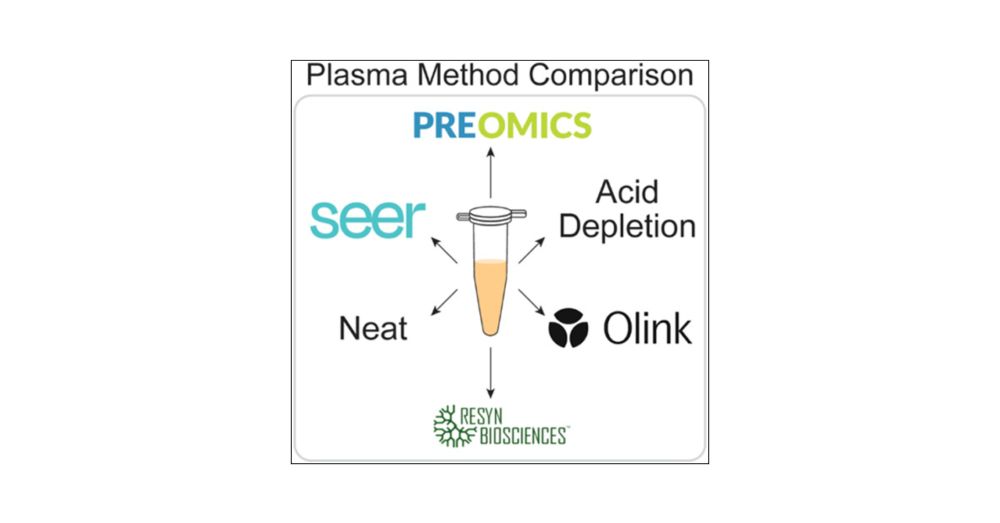
Technical Evaluation of Plasma Proteomics Technologies
Plasma proteomics technologies are rapidly evolving and of critical importance to the field of biomedical research. Here, we report a technical evaluation of six notable plasma proteomics technologies...
pubs.acs.org
Reposted by William Beimers
William Beimers
@diomed.bsky.social
· Jan 15
William Beimers
@diomed.bsky.social
· Jan 15
William Beimers
@diomed.bsky.social
· Jan 15
William Beimers
@diomed.bsky.social
· Jan 15
William Beimers
@diomed.bsky.social
· Jan 15

A Technical Evaluation of Plasma Proteomics Technologies
Plasma proteomic technologies are rapidly evolving and of critical importance to the field of biomedical research. Here we report a technical evaluation of six notable plasma proteomic technologies - ...
www.biorxiv.org
Reposted by William Beimers
Reposted by William Beimers
William Beimers
@diomed.bsky.social
· Nov 28
William Beimers
@diomed.bsky.social
· Nov 28

![[Person with ponytail wearing lab coat hands balloon to another person in front of a springboard, a magnet hanging from above, and then a target] PERSON WITH PONYTAIL to Person 2: Rub this balloon against your head, then go jump past that magnet toward the target on the wall. [caption] Before the bathroom scale was invented, the only way to weigh people was mass spectrometry.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:cz73r7iyiqn26upot4jtjdhk/bafkreid7lnvd7afmasx3nxi6anxmyhbatpaw54za3kiw3eaoclrf722faq@jpeg)



