
➡️ https://www.humangenetik.bio.lmu.de/research/index.html
Merry Christmas from the Hellnardos!

Merry Christmas from the Hellnardos!
In her thesis on the evolution of GRNs, she performed cross-species CRISPRi screens and helped establish primate iPSC lines.
Fiona was key for many social events and brought the Kölsch to Munich.
We will miss you and wish you all the best!

In her thesis on the evolution of GRNs, she performed cross-species CRISPRi screens and helped establish primate iPSC lines.
Fiona was key for many social events and brought the Kölsch to Munich.
We will miss you and wish you all the best!
We’re excited to welcome Jörg Beckmann (Nuremberg Zoo) for a talk on the new roles of modern zoos — from research to conservation and beyond.
🗓️ 12 Jan 2026, 4 PM
📍 LMU Biozentrum, Lecture Hall B01.019
See you there! 🦒🦍

We’re excited to welcome Jörg Beckmann (Nuremberg Zoo) for a talk on the new roles of modern zoos — from research to conservation and beyond.
🗓️ 12 Jan 2026, 4 PM
📍 LMU Biozentrum, Lecture Hall B01.019
See you there! 🦒🦍
👉 doi.org/10.1101/2025...

👉 doi.org/10.1101/2025...

He’s been a key part of the lab, contributing to many projects, including scRNAseq background and cross-species cell type comparisons, and always ready with new ideas (& coffee).
We’re sad to see him go but excited to see what comes next!



He’s been a key part of the lab, contributing to many projects, including scRNAseq background and cross-species cell type comparisons, and always ready with new ideas (& coffee).
We’re sad to see him go but excited to see what comes next!
He’ll present Cellbouncer, a new bioinformatic tool for pooled single-cell processing that yields insights into hominid evolution 🧬
More about the lab 👉 www.pollenlab.org

He’ll present Cellbouncer, a new bioinformatic tool for pooled single-cell processing that yields insights into hominid evolution 🧬
More about the lab 👉 www.pollenlab.org

We improved one of the most cost-efficient bulk RNAseq protocols out there to end up with +60% usable reads.
Check it out: doi.org/10.1101/2025...
a🧵1/9
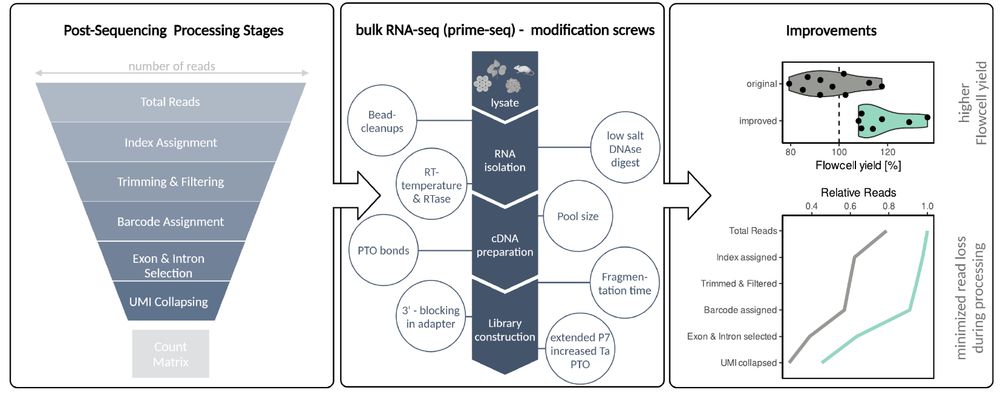
We improved one of the most cost-efficient bulk RNAseq protocols out there to end up with +60% usable reads.
Check it out: doi.org/10.1101/2025...
a🧵1/9
We introduce a systematic approach to maximize usable reads in RNA-seq.
With this, we created prime-seq2, improving one of the most cost-efficient bulk RNAseq protocols.
👉Check it out: doi.org/10.1101/2025...

We introduce a systematic approach to maximize usable reads in RNA-seq.
With this, we created prime-seq2, improving one of the most cost-efficient bulk RNAseq protocols.
👉Check it out: doi.org/10.1101/2025...
✨ Ines takes care of all the details that keep things organized
📑 Mrs Zhao is running the secretary
⛰️ Karin is mostly retired, but still comes in to help out
🧫 Sara is our greatest help in cell culture & ordering
Together, they make everything work smoothly 💙
✨ Ines takes care of all the details that keep things organized
📑 Mrs Zhao is running the secretary
⛰️ Karin is mostly retired, but still comes in to help out
🧫 Sara is our greatest help in cell culture & ordering
Together, they make everything work smoothly 💙
We are handling dogs, budgets, people, projects and databases.
Ines with Elly is running the dry lab
Wolfgang with Mo is running the wet lab
It’s such a privilege to work with all these wonderful people ❤️ and we have the best jobs we can imagine.
We are handling dogs, budgets, people, projects and databases.
Ines with Elly is running the dry lab
Wolfgang with Mo is running the wet lab
It’s such a privilege to work with all these wonderful people ❤️ and we have the best jobs we can imagine.
The „Wandertag“ of the Hellnardos in escape rooms!
Can you guess who was in which room?



The „Wandertag“ of the Hellnardos in escape rooms!
Can you guess who was in which room?
We are the single cell squad (scQUAD) 🙌
⛓️ Dana links peaks to genes
🕸️ Anita compares gene regulatory networks across primates
🎛️ Pascal tunes conservation metrics

We are the single cell squad (scQUAD) 🙌
⛓️ Dana links peaks to genes
🕸️ Anita compares gene regulatory networks across primates
🎛️ Pascal tunes conservation metrics
Welcome to the Pee-to-Profile Office!
🧪 Johanna developed a protocol to reprogram iPSCs from primate urine cells,
🧠 Manqi uses them to make neurons to study FOXP2,
🧫 Eva differentiates them to macrophages for cross-species comparisons and
📈 Daniel extends prime-seq to profile them all!

Welcome to the Pee-to-Profile Office!
🧪 Johanna developed a protocol to reprogram iPSCs from primate urine cells,
🧠 Manqi uses them to make neurons to study FOXP2,
🧫 Eva differentiates them to macrophages for cross-species comparisons and
📈 Daniel extends prime-seq to profile them all!
Meet the office of Leo, Antonia with Wilma and Felix
Leo 🧠 maps FOXP2 brains
Antonia 🧫 reprograms cross-species cells
Felix ⚙️ drives Part 2 of every project
Wilma 🐶 heads cable-integrity testing by chewing
#LabLife #BlueskyScience

Meet the office of Leo, Antonia with Wilma and Felix
Leo 🧠 maps FOXP2 brains
Antonia 🧫 reprograms cross-species cells
Felix ⚙️ drives Part 2 of every project
Wilma 🐶 heads cable-integrity testing by chewing
#LabLife #BlueskyScience

📃 doi.org/10.7554/eLif...
We investigated orthologous cell types in primate EBs and found that marker genes don't transfer well across species.
Check it out for insights into cross-species scRNA-seq and explore the data in our shiny app:
🔎 shiny.bio.lmu.de/Cross_Specie...
📃 doi.org/10.7554/eLif...
We investigated orthologous cell types in primate EBs and found that marker genes don't transfer well across species.
Check it out for insights into cross-species scRNA-seq and explore the data in our shiny app:
🔎 shiny.bio.lmu.de/Cross_Specie...
Here’s what we discovered 👇
www.biorxiv.org/content/10.1...

Here’s what we discovered 👇
www.biorxiv.org/content/10.1...
We are looking forward to reconnecting with everyone here and to welcoming new followers.
Let’s keep up great science interactions online!
We are looking forward to reconnecting with everyone here and to welcoming new followers.
Let’s keep up great science interactions online!

