
PI: @iAmphioxus.bsky.social
Web: www.evomicslab.org
targeting on genome instability evolution of both yeast and human genomes. Both computational genomics and experimental genomics backgrounds are welcome! 🧬🧑💻🧪 For more information, see evomicslab.org or send me a message!


www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
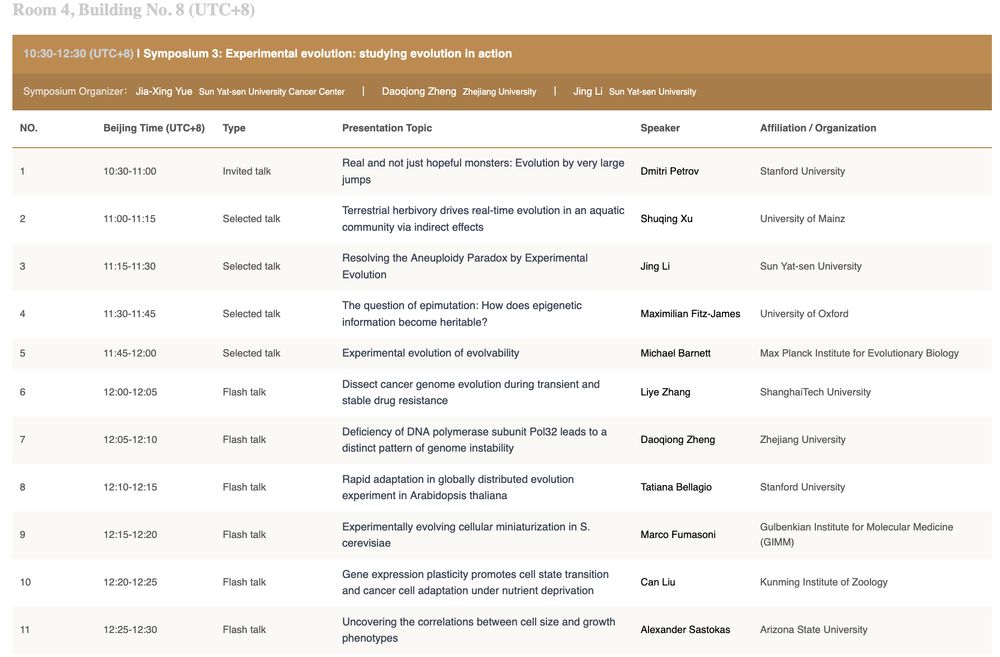
🔗 smbe2025.scimeeting.cn/en/web/program/25070


targeting on genome instability evolution of both yeast and human genomes. Both computational genomics and experimental genomics backgrounds are welcome! 🧬🧑💻🧪 For more information, see evomicslab.org or send me a message!
targeting on genome instability evolution of both yeast and human genomes. Both computational genomics and experimental genomics backgrounds are welcome! 🧬🧑💻🧪 For more information, see evomicslab.org or send me a message!
genome.cshlp.org/content/35/2...

genome.cshlp.org/content/35/2...
#LTEE #ExperimentalEvolution
We also invite you to register at www.smbe2025.org
@SMBEmeetings

#LTEE #ExperimentalEvolution
genome.cshlp.org/content/earl...
genome.cshlp.org/content/earl...
annual meeting in #Beijing .
Where: smbe2025.scimeeting.cn/en/web/index...
When: now - 2025/01/30. Welcome for all abstract submissions, especially for our Experimental Evolution Symposium !!!


annual meeting in #Beijing .
Where: smbe2025.scimeeting.cn/en/web/index...
When: now - 2025/01/30. Welcome for all abstract submissions, especially for our Experimental Evolution Symposium !!!
is open now!
Submission site: smbe2025.scimeeting.cn/en/web/index...
Submission Deadline: 2025.01.30
Welcome for all abstract submissions, especially for our Experimental Evolution Symposium !!!



is open now!
Submission site: smbe2025.scimeeting.cn/en/web/index...
Submission Deadline: 2025.01.30
Welcome for all abstract submissions, especially for our Experimental Evolution Symposium !!!




