🧬Code and notebooks will be released by the end of this week.
🎧Golden- Kpop Demon Hunters
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)

Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
More information here: europeanrosettacon.org
More information here: europeanrosettacon.org
rdcu.be/em0vA
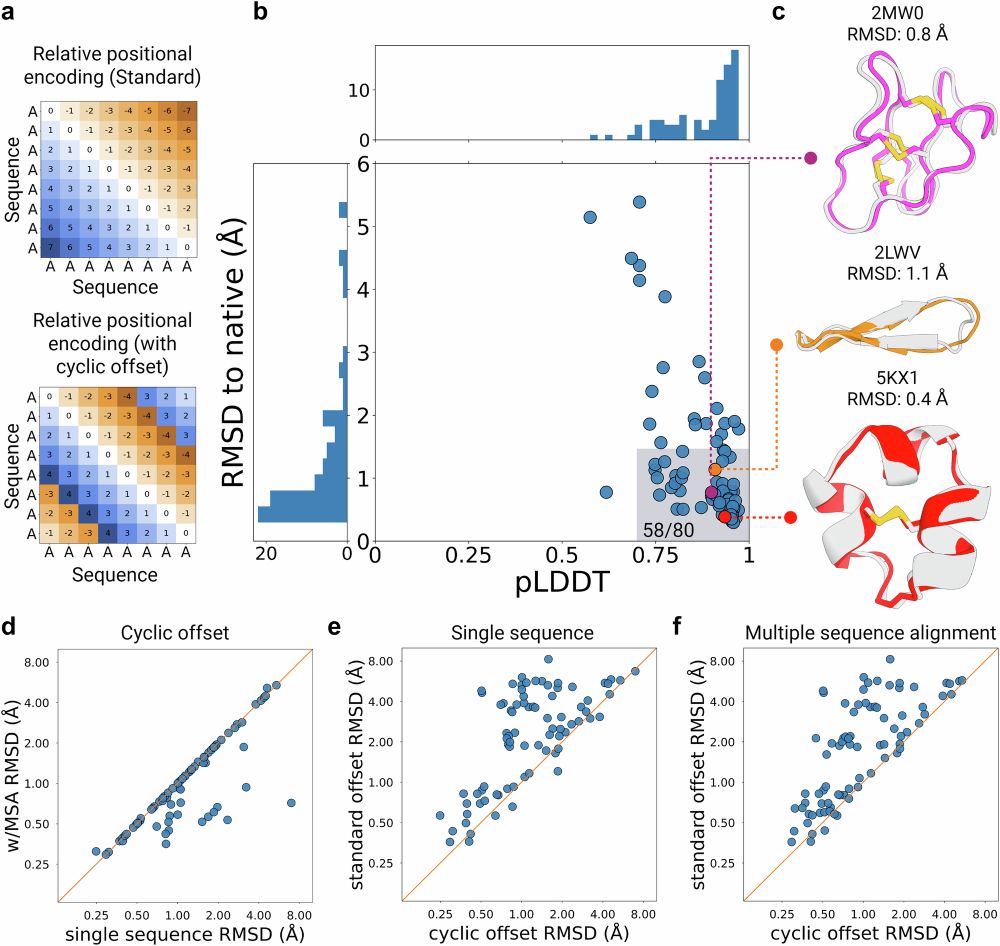
rdcu.be/em0vA
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
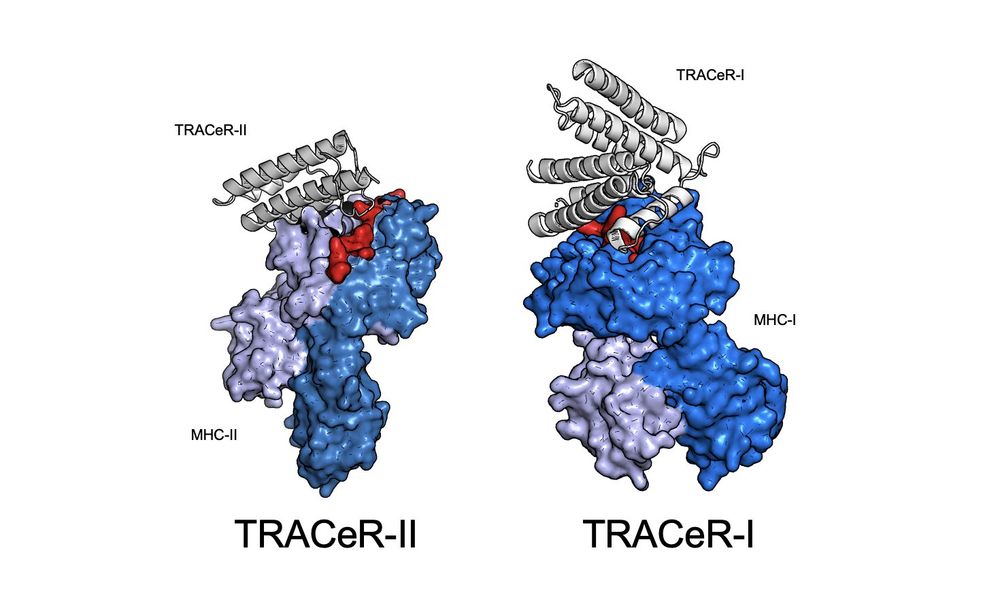
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
This week is packed with inspiration, press conferences and lectures, so stay tuned! 🌟
@uofwa.bsky.social @hhmi.bsky.social
#Science #AcademicSky

This week is packed with inspiration, press conferences and lectures, so stay tuned! 🌟
@uofwa.bsky.social @hhmi.bsky.social
#Science #AcademicSky
Preprint link: www.biorxiv.org/content/10.1...
Preprint link: www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

