
Opinions are my own.
@kdidi.bsky.social @machine.learning.bio @karstenkreis.bsky.social @arashv.bsky.social
arxiv.org/html/2507.09...

@kdidi.bsky.social @machine.learning.bio @karstenkreis.bsky.social @arashv.bsky.social
arxiv.org/html/2507.09...
📜 With wonderful co-authors, I'm co-presenting 4 main conference papers and 3
@gembioworkshop.bsky.social papers (gembio.ai), and I contribute to a panel (synthetic-data-iclr.github.io).
🧵 Overview in thread.
(1/n)

📜 With wonderful co-authors, I'm co-presenting 4 main conference papers and 3
@gembioworkshop.bsky.social papers (gembio.ai), and I contribute to a panel (synthetic-data-iclr.github.io).
🧵 Overview in thread.
(1/n)
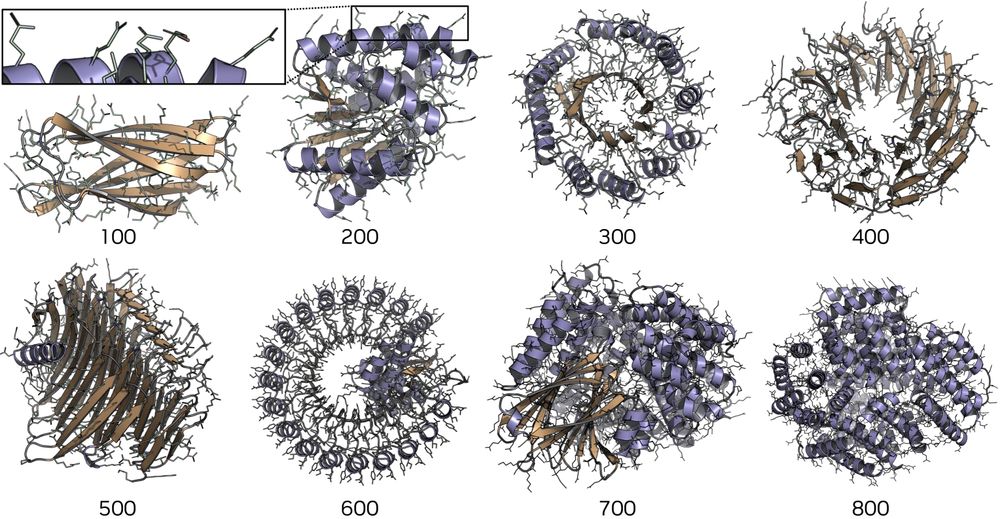
(i) Manually create new protein structure layouts? ✅
(ii) Generation with favorable designability/diversity/novelty trade-offs? ✅
(iii) Spatially edit given proteins? ✅
Very original work by the amazing @hannes-stark.bsky.social and Bowen Jing!🔥
ProtComposer: Compositional Protein Structure Generation with 3D Ellipsoids arxiv.org/abs/2503.05025
Condition on your 3D layout (of ellipsoids) to generate proteins like this or to get better designability/diversity/novelty tradeoffs.
1/6
(i) Manually create new protein structure layouts? ✅
(ii) Generation with favorable designability/diversity/novelty trade-offs? ✅
(iii) Spatially edit given proteins? ✅
Very original work by the amazing @hannes-stark.bsky.social and Bowen Jing!🔥
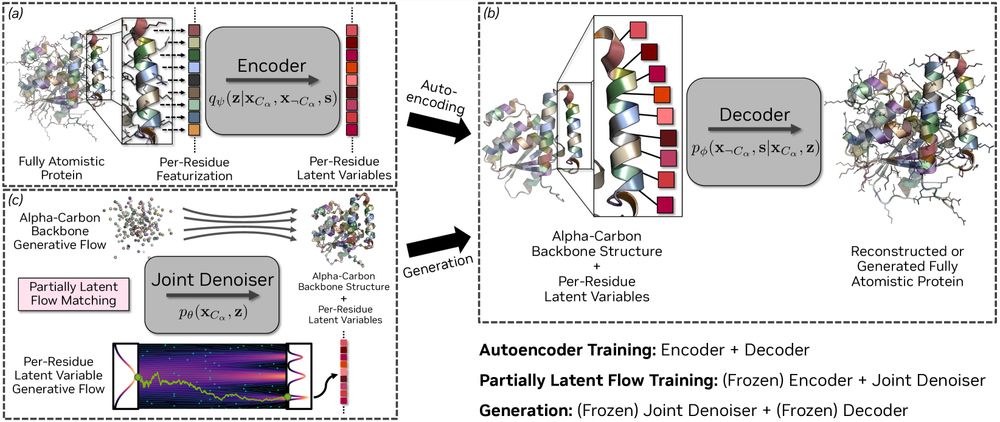
#ICLR2025 (Oral Presentation)
🔥 Project page: research.nvidia.com/labs/genair/...
📜 Paper: arxiv.org/abs/2503.00710
🛠️ Code and weights: github.com/NVIDIA-Digit...
🧵Details in thread...
(1/n)
#ICLR2025 (Oral Presentation)
🔥 Project page: research.nvidia.com/labs/genair/...
📜 Paper: arxiv.org/abs/2503.00710
🛠️ Code and weights: github.com/NVIDIA-Digit...
🧵Details in thread...
(1/n)

