
https://researchmap.jp/satoshi_kawato?lang=en
You can now display gene labels ONLY on the first genome in the linear diagram. Just select "First" in the "Label layout" option.
Try it out here👉: gbdraw.streamlit.app
#gbdraw #bioinformatics #genomics #visualization #virology #microbiology #streamlit


You can now display gene labels ONLY on the first genome in the linear diagram. Just select "First" in the "Label layout" option.
Try it out here👉: gbdraw.streamlit.app
#gbdraw #bioinformatics #genomics #visualization #virology #microbiology #streamlit
We reflect on how data explosion, new structural/AI tools, and complex features impact how we think about orthology link.springer.com/article/10.1...

We reflect on how data explosion, new structural/AI tools, and complex features impact how we think about orthology link.springer.com/article/10.1...
I also posted some recipes for viral genome diagrams on the GitHub Discussion thread. Hope you find them useful: github.com/satoshikawat...
#gbdraw #bioinformatics #genomics #visualization #virology #microbiology



I also posted some recipes for viral genome diagrams on the GitHub Discussion thread. Hope you find them useful: github.com/satoshikawat...
#gbdraw #bioinformatics #genomics #visualization #virology #microbiology
We show how phylogeny-based methods can resolve the problem of non-independence in genomic datasets.
These methods must be considered an essential part of the comparative genomics toolkit.
@lauriebelch.bsky.social @stuwest.bsky.social
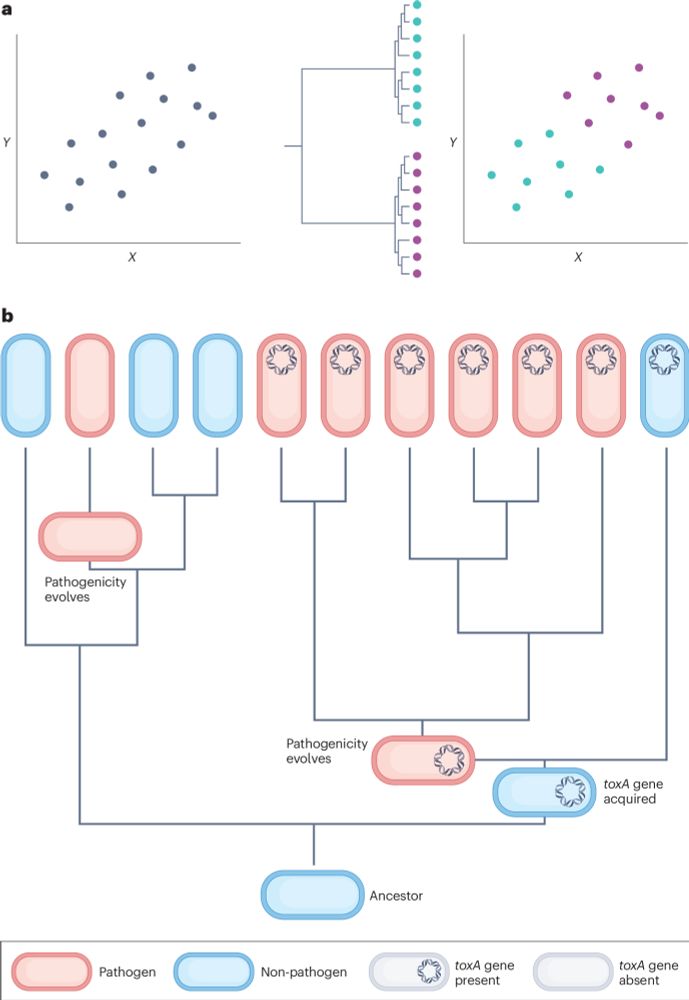
We show how phylogeny-based methods can resolve the problem of non-independence in genomic datasets.
These methods must be considered an essential part of the comparative genomics toolkit.
@lauriebelch.bsky.social @stuwest.bsky.social
- Feature overlap resolution
- Gradated BLAST hit ribbons
- Ruler-style scale
- Top/bottom legend placement
- Optimized defaults (for small genomes)
Try out: 👉 gbdraw.streamlit.app
#gbdraw #bioinformatics #genomics #visualization #virology

- Feature overlap resolution
- Gradated BLAST hit ribbons
- Ruler-style scale
- Top/bottom legend placement
- Optimized defaults (for small genomes)
Try out: 👉 gbdraw.streamlit.app
#gbdraw #bioinformatics #genomics #visualization #virology
@mkrupovic.bsky.social @deemteam.bsky.social @anagtz.bsky.social
www.nature.com/articles/s41...

@mkrupovic.bsky.social @deemteam.bsky.social @anagtz.bsky.social
www.nature.com/articles/s41...
- Added dinucleotide skew plotting to linear mode. Fixed the bug where only GC skew could be drawn.
- Added links to relevant sections of the documentation throughout the web app help menu.
Try out:👉 gbdraw.streamlit.app

- Added dinucleotide skew plotting to linear mode. Fixed the bug where only GC skew could be drawn.
- Added links to relevant sections of the documentation throughout the web app help menu.
Try out:👉 gbdraw.streamlit.app
- Smarter layout engine: reduced overlapping labels, legends, or definitions (linear mode)
- Circular mode: genomes >5 Mb now show tick labels in megabases (Mb)
- Official Documentation published with full guides & tutorials
🔗 gbdraw.streamlit.app

- Smarter layout engine: reduced overlapping labels, legends, or definitions (linear mode)
- Circular mode: genomes >5 Mb now show tick labels in megabases (Mb)
- Official Documentation published with full guides & tutorials
🔗 gbdraw.streamlit.app
Thread 1/n

Thread 1/n
gbdrawは、ブラウザ上で環状・直鎖ゲノムを描画できるウェブアプリ (ローカルCUI版あり) です。論文や発表の図作成にぜひご活用ください。
gbdraw.streamlit.app
github.com/satoshikawat...

gbdrawは、ブラウザ上で環状・直鎖ゲノムを描画できるウェブアプリ (ローカルCUI版あり) です。論文や発表の図作成にぜひご活用ください。
gbdraw.streamlit.app
github.com/satoshikawat...
I’ve completely revamped GC content/skew calculations and label processing, resulting in a dramatic speed boost.
Install via Bioconda, or try it on the web app!
github.com/satoshikawat...
I’ve completely revamped GC content/skew calculations and label processing, resulting in a dramatic speed boost.
Install via Bioconda, or try it on the web app!
github.com/satoshikawat...
Try out: gbdraw.streamlit.app

Try out: gbdraw.streamlit.app
- Support for GFF3+FASTA format input
- New label whitelist feature
- Local GUI version (optional; requires extra installation of Streamlit)
Please try our web app version until the Bioconda release is complete.
Check it out! gbdraw.streamlit.app

- Support for GFF3+FASTA format input
- New label whitelist feature
- Local GUI version (optional; requires extra installation of Streamlit)
Please try our web app version until the Bioconda release is complete.
Check it out! gbdraw.streamlit.app
- Label blacklist
- Custom label qualifiers
- Adjustable label arcs & font size
- Configurable axis color & width
Install with mamba or try online: gbdraw.streamlit.app


- Label blacklist
- Custom label qualifiers
- Adjustable label arcs & font size
- Configurable axis color & width
Install with mamba or try online: gbdraw.streamlit.app
- selectively hide labels via a blacklist
- qualifier display priority customization
- adjustable label arcs
These updates make it easier to visualize larger genomes without layout issues.
gbdraw.streamlit.app




- selectively hide labels via a blacklist
- qualifier display priority customization
- adjustable label arcs
These updates make it easier to visualize larger genomes without layout issues.
gbdraw.streamlit.app
- You can now place labels on the inner edge of circular genomes.
- Edit the color palette directly within the app.
👉Try it out: gbdraw.streamlit.app




- You can now place labels on the inner edge of circular genomes.
- Edit the color palette directly within the app.
👉Try it out: gbdraw.streamlit.app
gbdraw.streamlit.app


gbdraw.streamlit.app
blast2dotplot.streamlit.app

blast2dotplot.streamlit.app
skewplot.streamlit.app

skewplot.streamlit.app
orfind.streamlit.app

orfind.streamlit.app
Google Colab version also available:
github.com/satoshikawat...

Google Colab version also available:
github.com/satoshikawat...
Visualize annotated genomes with a single command.
github.com/satoshikawat...

Visualize annotated genomes with a single command.
github.com/satoshikawat...
colab.research.google.com/github/satos...


colab.research.google.com/github/satos...

