
go.nature.com/4pBArFH

go.nature.com/4pBArFH
You can now back the campaign to get your very own pre-assembled Game Bub!
www.crowdsupply.com/second-bedro...

You can now back the campaign to get your very own pre-assembled Game Bub!
www.crowdsupply.com/second-bedro...
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵

Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Sign up for updates and to hear when the campaign goes live: www.crowdsupply.com/second-bedro...
This will be an assembled product, not just a kit!

Sign up for updates and to hear when the campaign goes live: www.crowdsupply.com/second-bedro...
This will be an assembled product, not just a kit!


www.washingtonpost.com/health/2025/...

www.washingtonpost.com/health/2025/...


I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
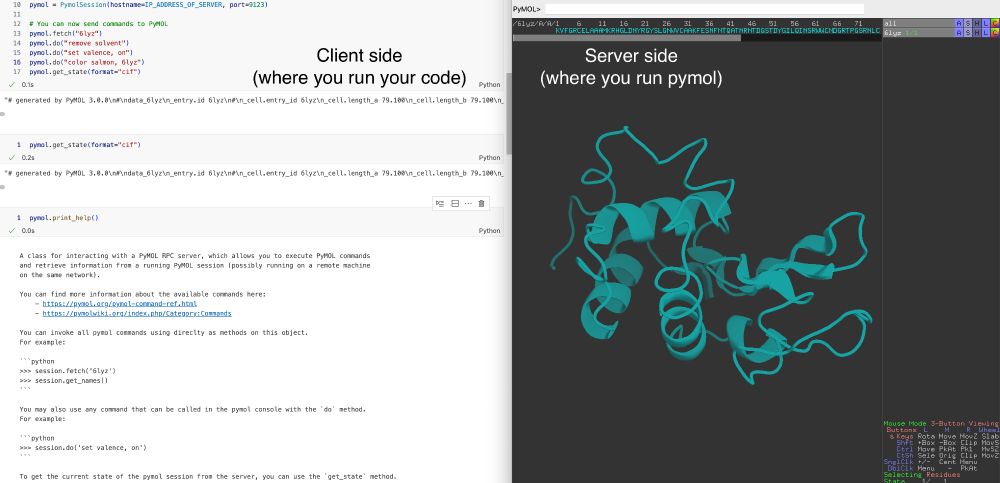
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
RLA is a contrastive-learning approach that assesses sequence-structure compatibility by aligning sequence and structure machine learning representations! RLA can successfully filter protein binder designs. 1/3 🧶
journals.aps.org/prxlife/abst...

RLA is a contrastive-learning approach that assesses sequence-structure compatibility by aligning sequence and structure machine learning representations! RLA can successfully filter protein binder designs. 1/3 🧶
journals.aps.org/prxlife/abst...

