www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...

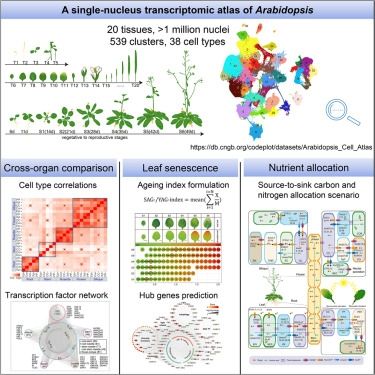
www.biorxiv.org/content/10.1...
Here we show that drought response supersedes differences in cell identity and identify key transcriptional regulators.

www.biorxiv.org/content/10.1...
Here we show that drought response supersedes differences in cell identity and identify key transcriptional regulators.
nph.onlinelibrary.wiley.com/doi/10.1111/...
"How evolution repeatedly builds complexity: a case study with C4 photosynthesis in Blepharis (Acanthaceae)"
And this was chosen for a cover photo:

nph.onlinelibrary.wiley.com/doi/10.1111/...
"How evolution repeatedly builds complexity: a case study with C4 photosynthesis in Blepharis (Acanthaceae)"
And this was chosen for a cover photo:
To Learn More, Visit: www.psna2025.com
@psna-official.bsky.social

To Learn More, Visit: www.psna2025.com
@psna-official.bsky.social
Available here: onlinelibrary.wiley.com/doi/10.1002/...

Available here: onlinelibrary.wiley.com/doi/10.1002/...

UnigeneFinder is a software tool designed to reduce the redundancy of de novo transcriptome assemblies and produce sets of primary transcripts, coding sequences, and proteins, along with gene expression data.
dx.doi.org/10.1002/pld3...

UnigeneFinder is a software tool designed to reduce the redundancy of de novo transcriptome assemblies and produce sets of primary transcripts, coding sequences, and proteins, along with gene expression data.
dx.doi.org/10.1002/pld3...
Monson et al.
👇
📖 nph.onlinelibrary.wiley.com/doi/full/10....
#LatestIssue
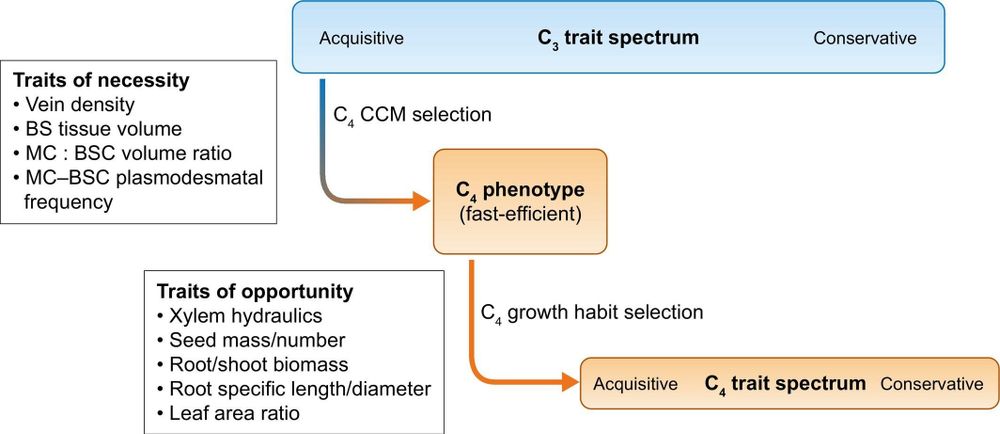
Monson et al.
👇
📖 nph.onlinelibrary.wiley.com/doi/full/10....
#LatestIssue

Early Bird Discount deadline is April 20!
To Register: www.psna2025.com
@psna-official.bsky.social

Early Bird Discount deadline is April 20!
To Register: www.psna2025.com
@psna-official.bsky.social