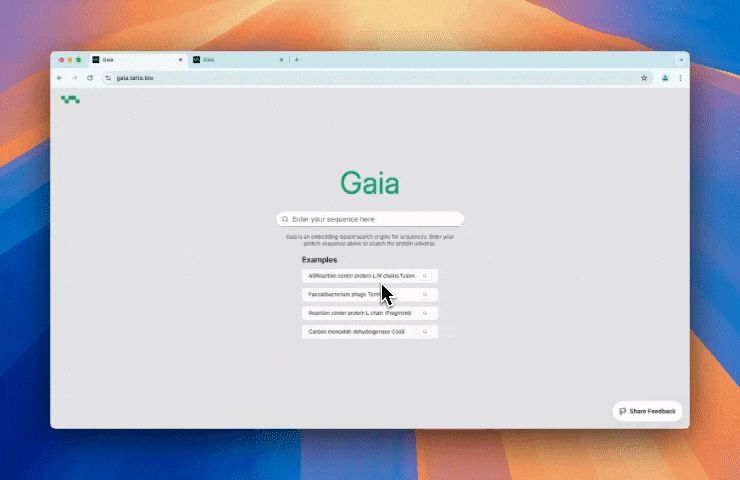
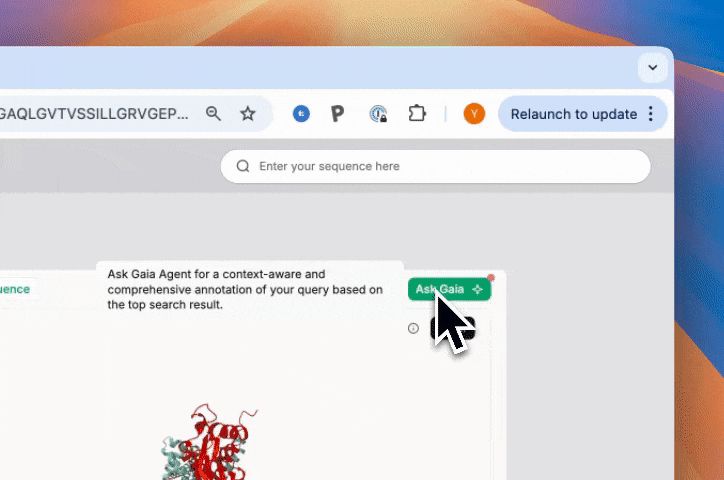
Over 1,000 scientists worldwide have already used SeqHub to annotate more than 550,000 proteins, uncovering new insights and accelerating discovery.
Over 1,000 scientists worldwide have already used SeqHub to annotate more than 550,000 proteins, uncovering new insights and accelerating discovery.
Registration for our 2025 NeLLi Symposium is now open. For the first time in collaboration with @unlv.edu
Mark the date: November 6-7 in Las Vegas, NV
Join us in November for talks focused on the most recent expansions of the Tree of Life, and the latest discoveries toward the evolution of cellular complexity and microbial symbiosis.
Learn more: jointgeno.me/NeLLi
@nigelmouncey.bsky.social

Registration for our 2025 NeLLi Symposium is now open. For the first time in collaboration with @unlv.edu
Mark the date: November 6-7 in Las Vegas, NV

www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵

www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵



www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
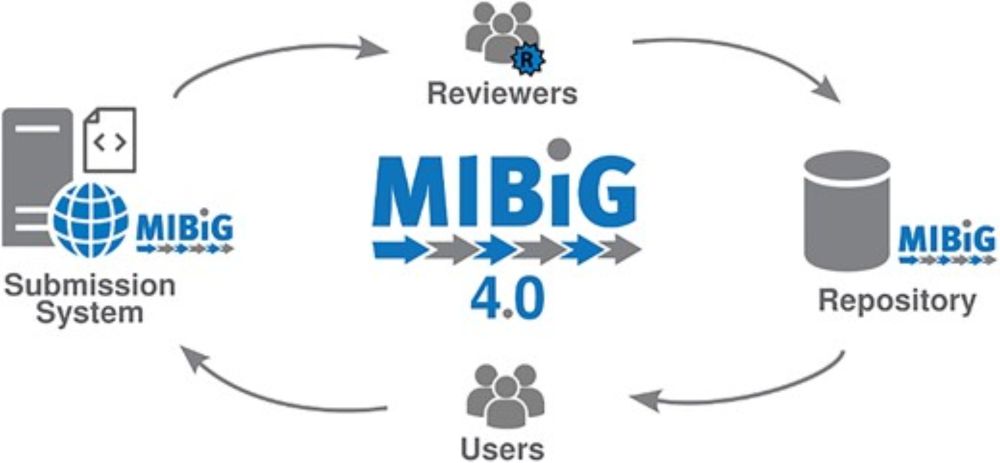
bit.ly/plaid-proteins
🧵
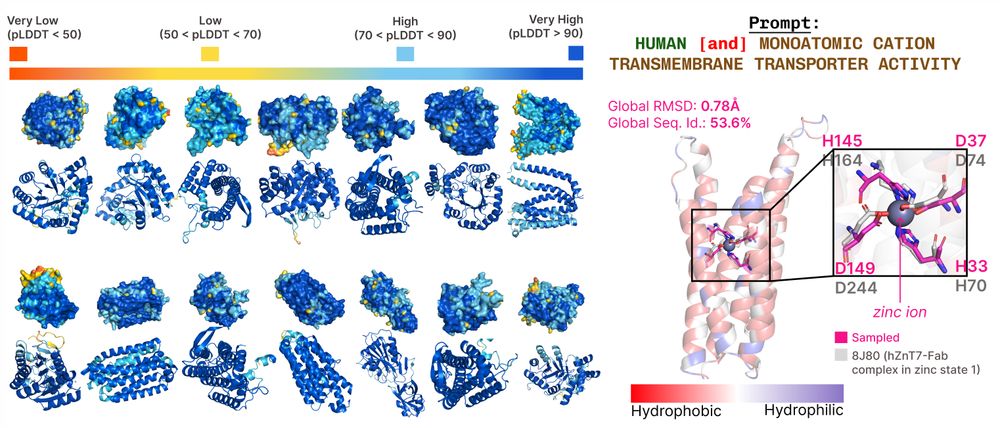
bit.ly/plaid-proteins
🧵
🌐 bfvd.foldseek.com
💾 bfvd.steineggerlab.workers.dev
📄 academic.oup.com/nar/advance-...
🌐 bfvd.foldseek.com
💾 bfvd.steineggerlab.workers.dev
📄 academic.oup.com/nar/advance-...