In new work @arcinstitute.org, we report the discovery and engineering of the first programmable DNA recombinases capable of megabase-scale human genome rearrangement
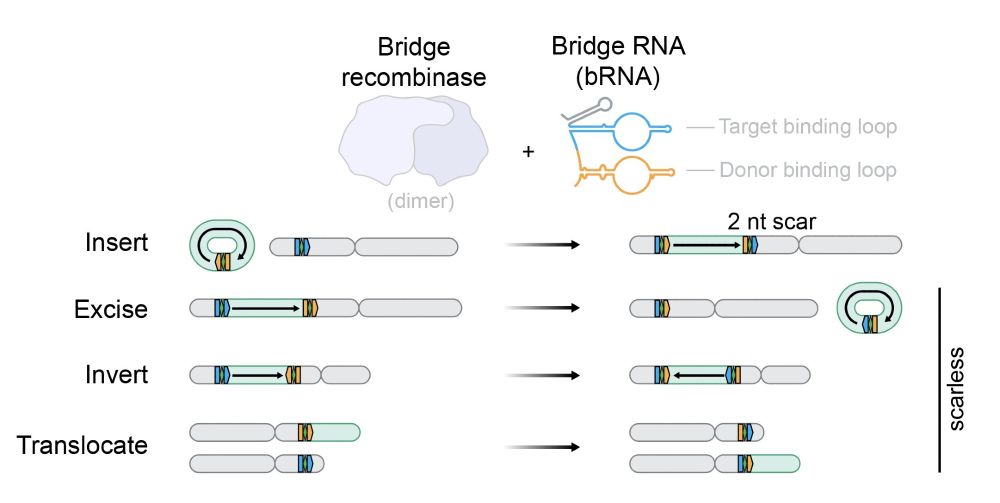
In new work @arcinstitute.org, we report the discovery and engineering of the first programmable DNA recombinases capable of megabase-scale human genome rearrangement
endpts.com/endpoints-sl...

endpts.com/endpoints-sl...
Today we launch scBaseCamp, the largest public repository of single cell RNAseq data, uniformly processed from raw sequencing reads.

Today we launch scBaseCamp, the largest public repository of single cell RNAseq data, uniformly processed from raw sequencing reads.

www.youtube.com/watch?v=ak_f...

www.youtube.com/watch?v=ak_f...
Evo 2 now includes information from all domains in life to expand its capabilities in generative functional genomics. @pdhsu.bsky.social @brianhie.bsky.social
tinyurl.com/3t83vseh

Evo 2 now includes information from all domains in life to expand its capabilities in generative functional genomics. @pdhsu.bsky.social @brianhie.bsky.social
tinyurl.com/3t83vseh
Today, @arcinstitute.org in collaboration with Nvidia releases Evo 2—a fully open source biological foundation model trained on genomes spanning the entire tree of life.
Today, @arcinstitute.org in collaboration with Nvidia releases Evo 2—a fully open source biological foundation model trained on genomes spanning the entire tree of life.