Also includes high quality muscle single cell data via GEO.
nature.com/articles/s43.... Bluetorial on #cachexia, #cancer, #cardiovascular health
@uicancercenter.bsky.social @uofilsystem.bsky.social
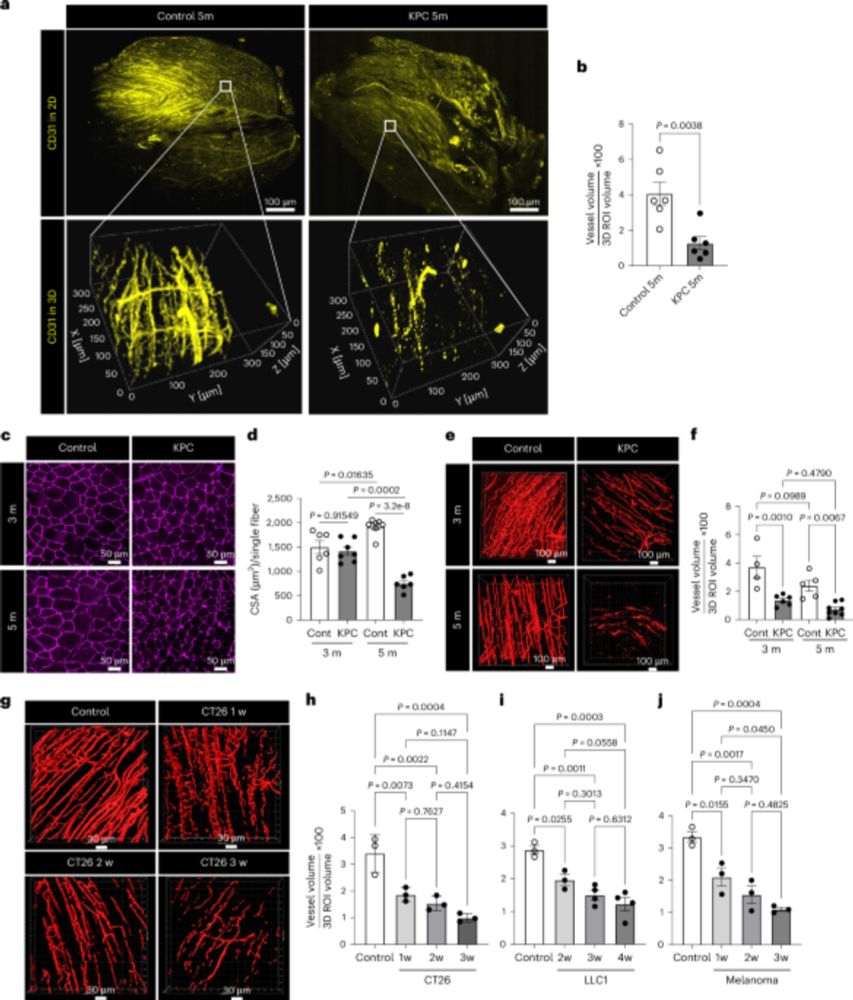
Also includes high quality muscle single cell data via GEO.
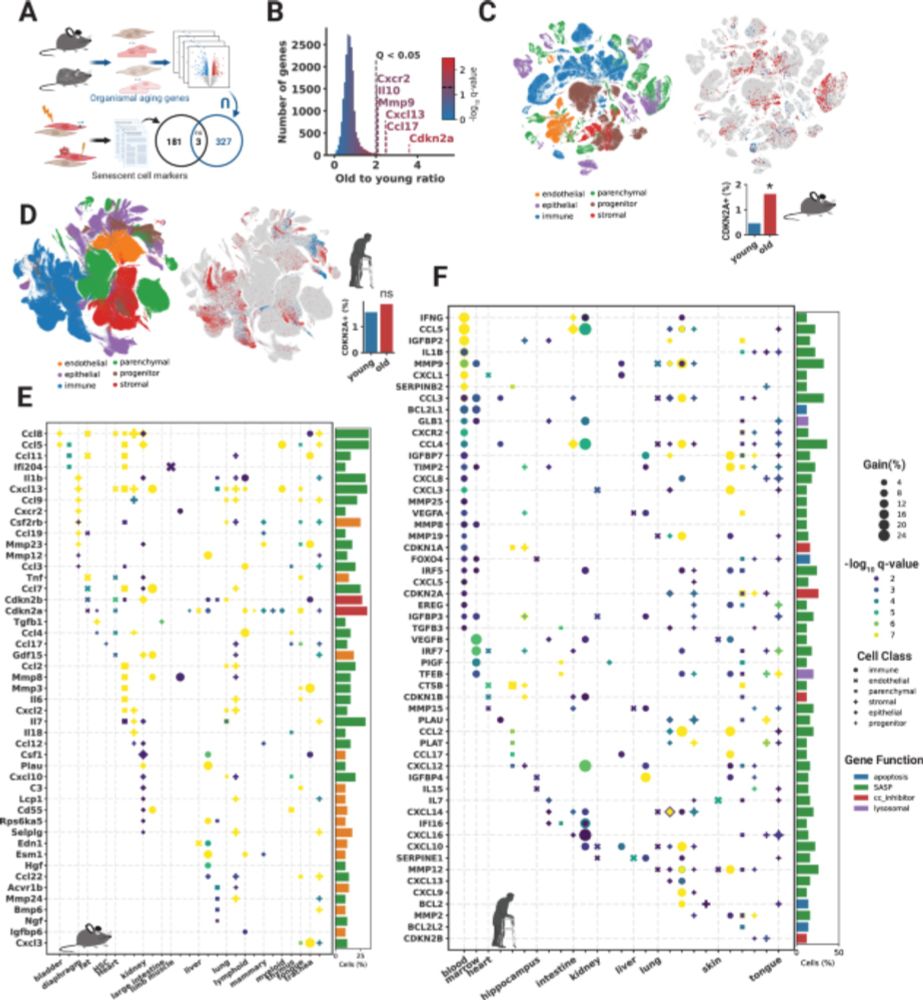
Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
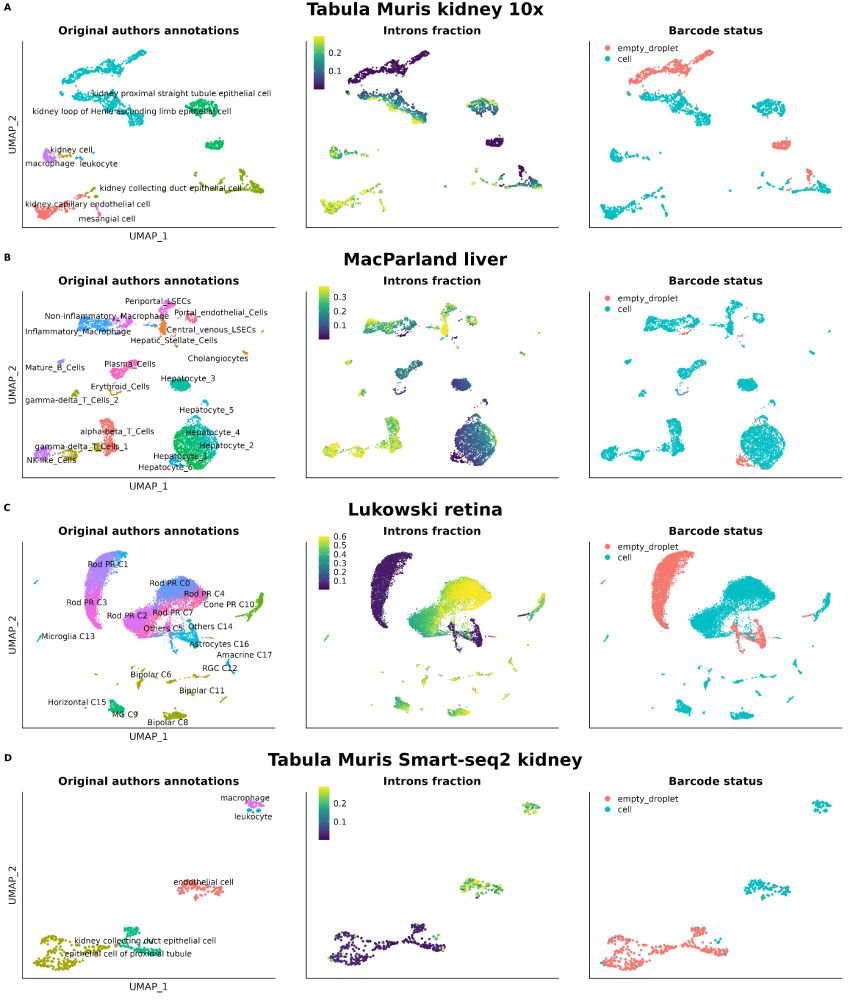
Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
I’ve made a short video covering this simple but important concept:
youtu.be/r4A_QgseUfw?...

I’ve made a short video covering this simple but important concept:
youtu.be/r4A_QgseUfw?...
I'm at the University of Colorado. I often say that if you pick two of three from #transcriptome, #ML, and #publicdata, my lab is probably interested.
I'm at the University of Colorado. I often say that if you pick two of three from #transcriptome, #ML, and #publicdata, my lab is probably interested.


