
Topic: Synergistic coevolution in mono-specific and multi-species microbial consortia
Please RT or forward this information to interested candidates.
Deadline: 11.01.26
More info:
shorturl.at/f1TuF

Topic: Synergistic coevolution in mono-specific and multi-species microbial consortia
Please RT or forward this information to interested candidates.
Deadline: 11.01.26
More info:
shorturl.at/f1TuF
www.biospektrum.de/system/files...
@dger.bsky.social

Work lead by the amazing Kevser Bilici!

Work lead by the amazing Kevser Bilici!

Work lead by the amazing Kevser Bilici!
www.findaphd.com/phds/project...

www.findaphd.com/phds/project...
So many people to thank... We are a fantastic team!
Our CMFI Cluster of Excellence @unituebingen.bsky.social receives funding extension for the next seven years.
Spokesperson Andreas Peschel @andreaspeschel.bsky.social: "We can now advance our research into resistance mechanisms and new antimicrobial agents!"
shorturl.at/rcK1A
So many people to thank... We are a fantastic team!

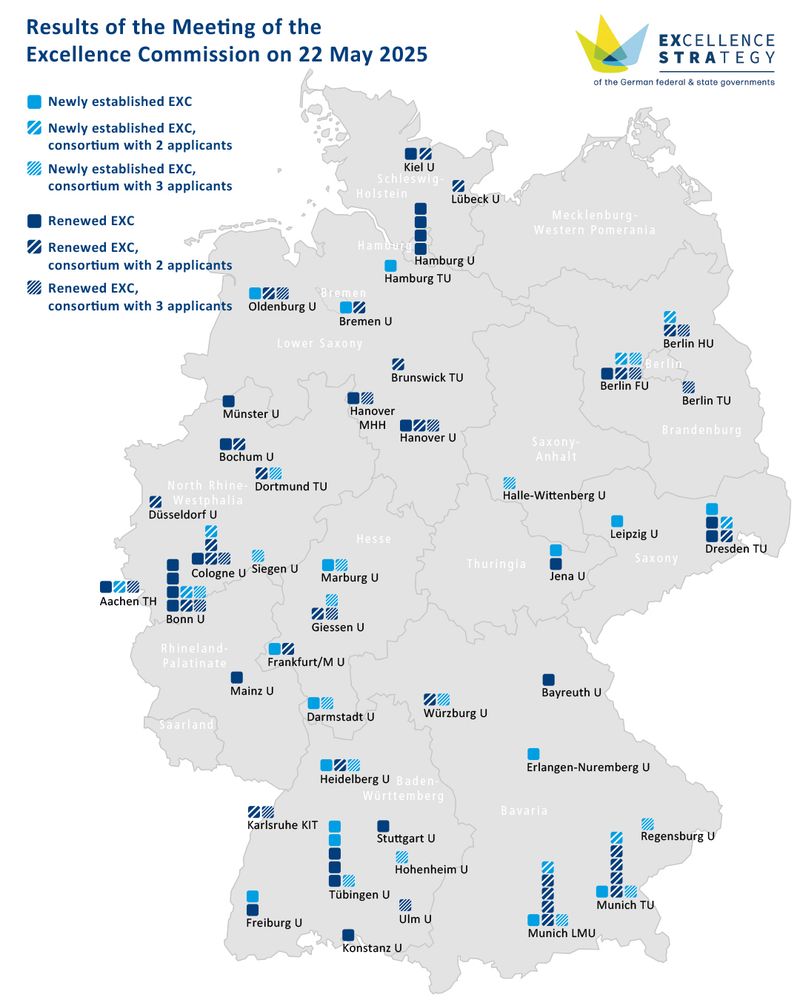
uni-tuebingen.de/universitaet...

uni-tuebingen.de/universitaet...
www.biorxiv.org/content/10.1...
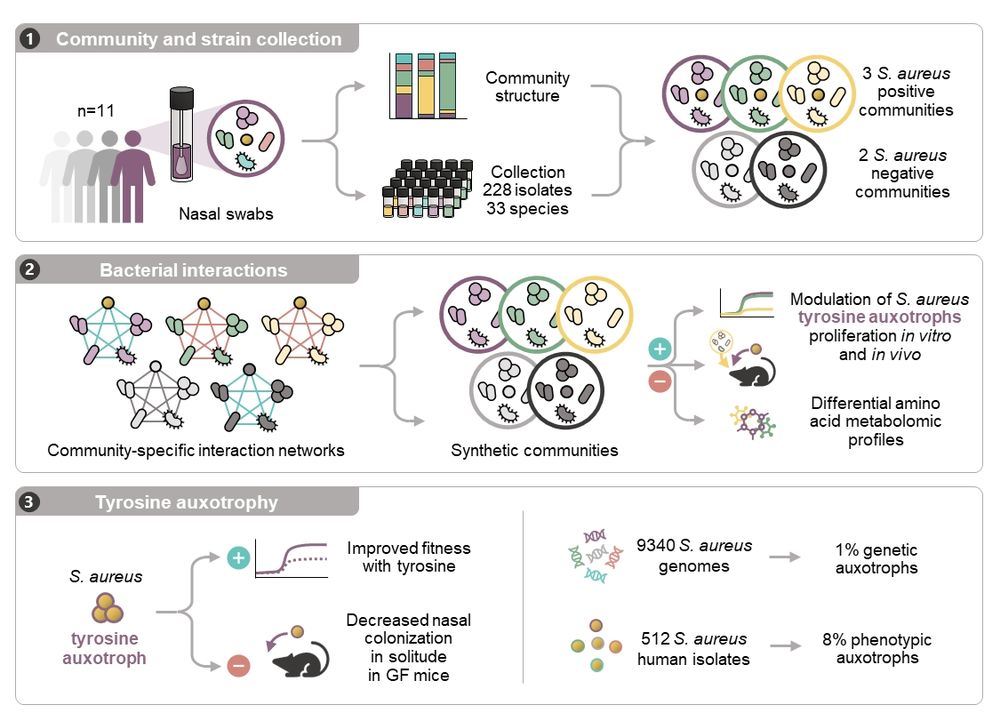
www.biorxiv.org/content/10.1...
www.sciencedirect.com/science/arti...
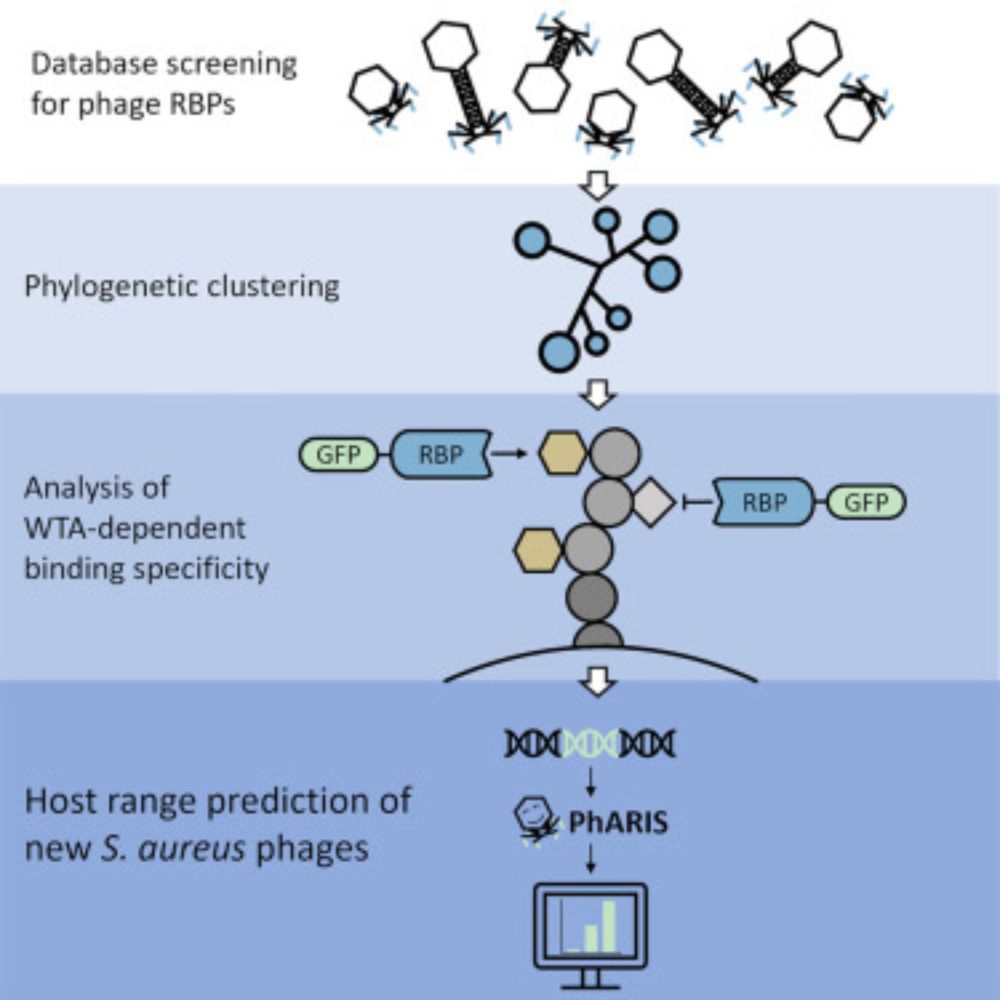
www.sciencedirect.com/science/arti...
www.science.org/doi/10.1126/...
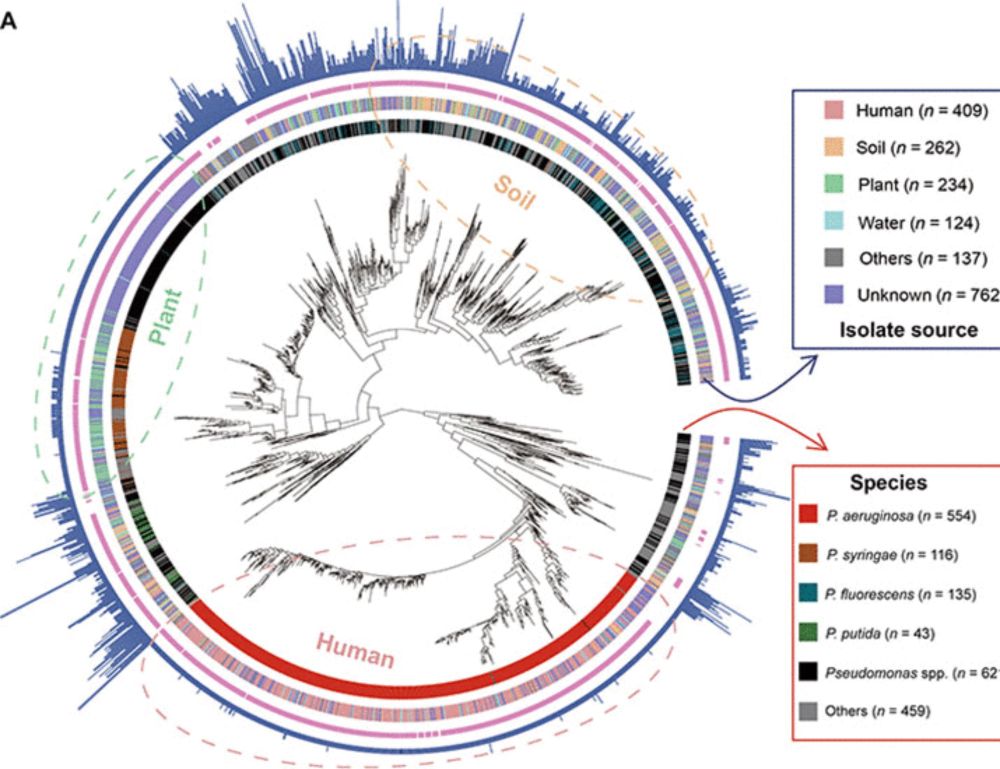
www.science.org/doi/10.1126/...
journals.asm.org/doi/10.1128/...


