Check it out here: www.biorxiv.org/content/10.1...

Rewatch the kickoff event: tinyurl.com/37cz9bnj

Rewatch the kickoff event: tinyurl.com/37cz9bnj
Speaking tonight at #ISMB2025 in the CompMS track about making proteomics AI ready. Expect metadata, MLMarker, and a lot of public data love. See you there!
@iscb.bsky.social
Speaking tonight at #ISMB2025 in the CompMS track about making proteomics AI ready. Expect metadata, MLMarker, and a lot of public data love. See you there!
@iscb.bsky.social
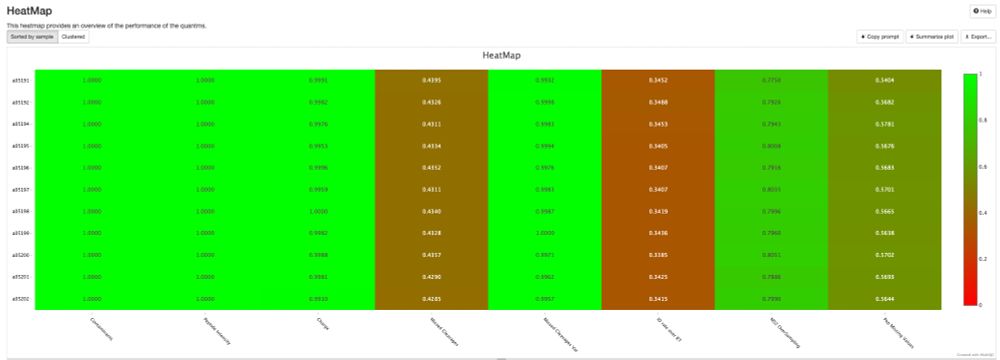
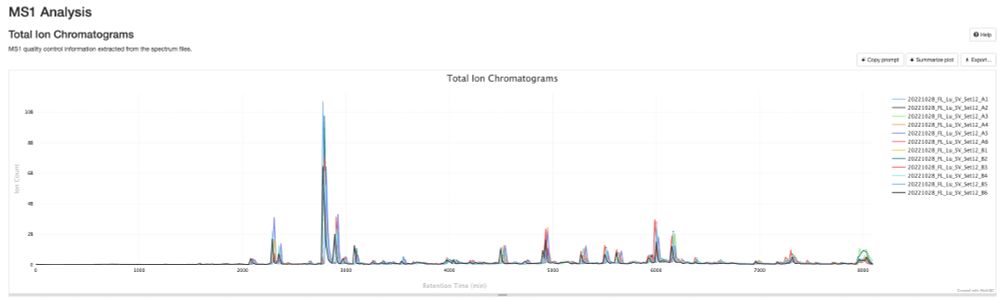
💡 Create stunning, shareable HTML reports for your collaborators in seconds.
✨ Try pmultiqc.quantms.org Examples👇
#Proteomics #QC
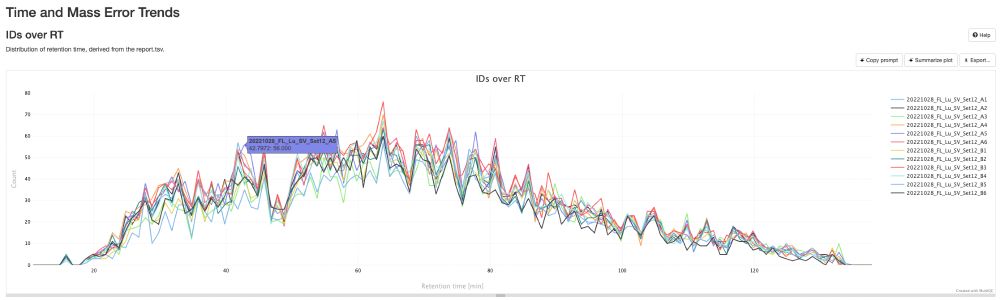
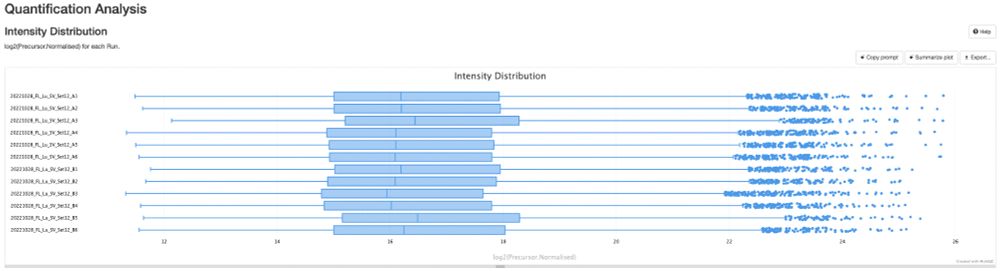
💡 Create stunning, shareable HTML reports for your collaborators in seconds.
✨ Try pmultiqc.quantms.org Examples👇
#Proteomics #QC
It's like the philosopher's stone, turning your experimental data into gold and giving it the elixir of life.
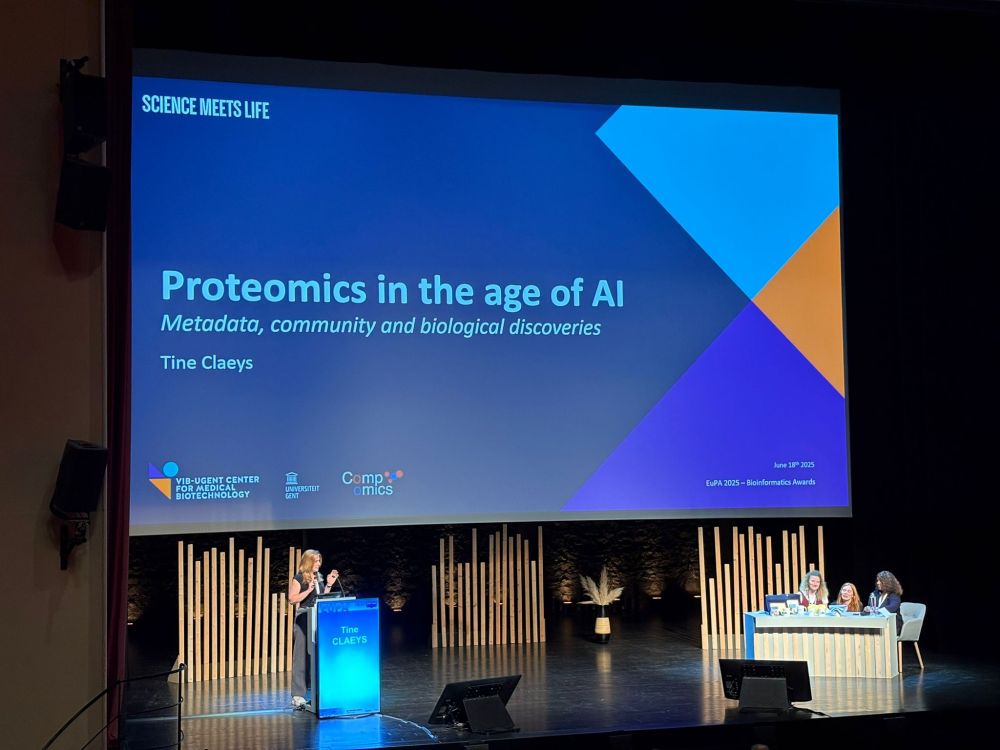
Congrats with the EuPA Bioinformatics Award, Tine!
#EuPA2025

It's like the philosopher's stone, turning your experimental data into gold and giving it the elixir of life.
Congrats with the EuPA Bioinformatics Award, Tine!
#EuPA2025
Take 15 mins to tell funders why open data matters!
📊 Fill out the EMBL-EBI 2025 survey 👉
www.surveymonkey.com/r/QGFMBH8?ch...
Your feedback helps keep PRIDE open, FAIR & impactful.
🙏 Please share!
#FAIR #OpenData #Proteomics #MassSpectrometry #PRIDE
Take 15 mins to tell funders why open data matters!
📊 Fill out the EMBL-EBI 2025 survey 👉
www.surveymonkey.com/r/QGFMBH8?ch...
Your feedback helps keep PRIDE open, FAIR & impactful.
🙏 Please share!
#FAIR #OpenData #Proteomics #MassSpectrometry #PRIDE
Preprint & app: www.biorxiv.org/content/10.1...
Let's chat at #EuPA2025 - Award session (Wednesday) & poster session (Thursday)!
Preprint & app: www.biorxiv.org/content/10.1...
Let's chat at #EuPA2025 - Award session (Wednesday) & poster session (Thursday)!
After a short introduction, we have three exciting talks lined up, as well as an interactive discussion on open issues in computational proteomics.
@eupaproteomics.bsky.social @uszkoreitju.bsky.social @harirmds.bsky.social

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
We’re officially in #ASMS2025 mode and kicking things off with a major milestone:
🚀 Introducing PRIDE-AP – the first Affinity Proteomics archive!
🔗 www.ebi.ac.uk/pride/archiv...
A new home for your #Olink and #SomaScan and other non-MS data.👇
We’re officially in #ASMS2025 mode and kicking things off with a major milestone:
🚀 Introducing PRIDE-AP – the first Affinity Proteomics archive!
🔗 www.ebi.ac.uk/pride/archiv...
A new home for your #Olink and #SomaScan and other non-MS data.👇
Now it is your turn! And it is such a great opportunity to support #ECRs and grow with the #proteomics community 🥰
Highly recommend applying! 🙌
🗓️ Apps open June 4 until July 2
📥 Apply here: forms.gle/t5jhWCmtxWUX...
📢 Be the change in science leadership!
#ScientificLeadership #BoardElections

Now it is your turn! And it is such a great opportunity to support #ECRs and grow with the #proteomics community 🥰
Highly recommend applying! 🙌

I’ve pushed a new version of lesSDRF to make high-throughput SDRF annotation faster and more intuitive.
Github issue: github.com/CompOmics/le...
Temporary app: refactorlessdrf.streamlit.app
I could use some feedback before making this the new lesSDRF 🙏
I’ve pushed a new version of lesSDRF to make high-throughput SDRF annotation faster and more intuitive.
Github issue: github.com/CompOmics/le...
Temporary app: refactorlessdrf.streamlit.app
I could use some feedback before making this the new lesSDRF 🙏

💡Juan Antonio @pride-ebi.bsky.social first talked about what really is AI-ready data, what is the end goal. (1/5)




💡Juan Antonio @pride-ebi.bsky.social first talked about what really is AI-ready data, what is the end goal. (1/5)
Get ahead of the curve—check them out & start your submissions! 👇
🔗 github.com/PRIDE-Archiv...
#Proteomics #Olink #SomaScan
Get ahead of the curve—check them out & start your submissions! 👇
🔗 github.com/PRIDE-Archiv...
#Proteomics #Olink #SomaScan
Check it out here:
www.biorxiv.org/content/10.1...

Check it out here:
www.biorxiv.org/content/10.1...

Our latest research, TIMS²Rescore, is now published in Journal of Proteome Research! 🎉
Read it here: pubs.acs.org/doi/full/10....
A huge thanks to all our collaborators for making this happen!

Our latest research, TIMS²Rescore, is now published in Journal of Proteome Research! 🎉
Read it here: pubs.acs.org/doi/full/10....
A huge thanks to all our collaborators for making this happen!
#EuBIC2025

#EuBIC2025