

Imagine looking at the distribution of the allelic coverage fraction at heterozygous calls in an indivdual and getting this distribution 👇 😱😱😱
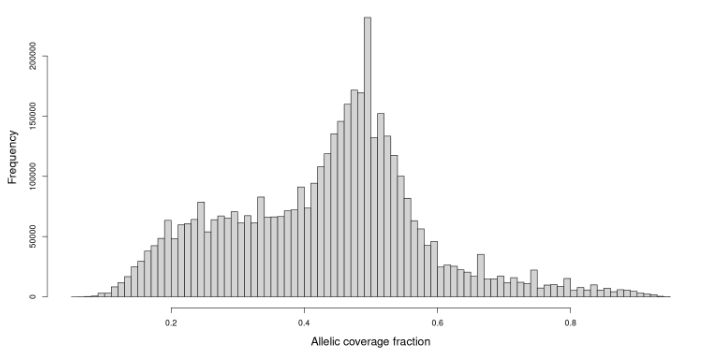
Imagine looking at the distribution of the allelic coverage fraction at heterozygous calls in an indivdual and getting this distribution 👇 😱😱😱
Do you like speciation? Genomics? Hybridization? Bioinformatics? Then this is for you:
Published today: "The Distribution & Dispersal of Large Haploblocks in a Superspecies"
Bonus interest if you like ring species and cute greenish birds:
doi.org/10.1111/mec....

Do you like speciation? Genomics? Hybridization? Bioinformatics? Then this is for you:
Published today: "The Distribution & Dispersal of Large Haploblocks in a Superspecies"
Bonus interest if you like ring species and cute greenish birds:
doi.org/10.1111/mec....
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...



In this study, SNP panels were designed for Lake Whitefish, Brook trout & Arctic Charr for usage in indigenous-led & commercial fisheries management initiatives, including mine 😎.
doi.org/10.1111/1755...

In this study, SNP panels were designed for Lake Whitefish, Brook trout & Arctic Charr for usage in indigenous-led & commercial fisheries management initiatives, including mine 😎.
doi.org/10.1111/1755...

