AIMD results came from: doi.org/10.1103/PRXE...
They also compared with Neutron Diffraction. @myzzzz.bsky.social will benchmark the S(Q) with experimental data.
#compchemsky #compchem


AIMD results came from: doi.org/10.1103/PRXE...
They also compared with Neutron Diffraction. @myzzzz.bsky.social will benchmark the S(Q) with experimental data.
#compchemsky #compchem

#compchem #compchemsky

#compchem #compchemsky
#compchem #compchemsky


#compchem #compchemsky
#CompChem #ChemChemSky

#CompChem #ChemChemSky

The newly released open-source model from OpenAI can be tried on TractoAI playground for *FREE*.

The newly released open-source model from OpenAI can be tried on TractoAI playground for *FREE*.
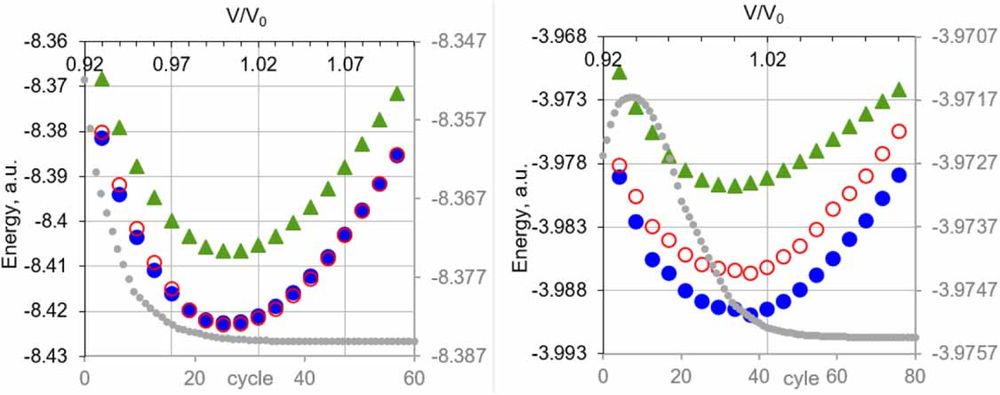


#CompChemSky #CompChem #GPU

#CompChemSky #CompChem #GPU




Inspired by www.youtube.com/watch?v=mDyt...
#compchem #compchemsky
Inspired by www.youtube.com/watch?v=mDyt...
#compchem #compchemsky
#compchemsky #compchem
colab.research.google.com/drive/1g1PiW...

#compchemsky #compchem
colab.research.google.com/drive/1g1PiW...
github.com/aligfellow/v...
Returns a list of internal coordinates associated with the vibrational mode, can be used from command line on xyz trjfiles or orca or gaussian outputs etc.

#compchemsky #chemsky #compchem



#compchemsky #chemsky #compchem
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...

Join us in Zoom: us06web.zoom.us/j/8607128794...
#compchem #dft

Join us in Zoom: us06web.zoom.us/j/8607128794...
#compchem #dft
#compchem #CompChemSky
#compchem #CompChemSky

#compchemsky #chemsky
#compchemsky #chemsky
open.substack.com/pub/quantabr...

open.substack.com/pub/quantabr...







