Website: http://abacus.gene.ucl.ac.uk/
BPP GitHub: https://github.com/bpp/
PAML GitHub: https://github.com/abacus-gene/paml
PAML discussion group: https://groups.google.com/g/pamlsoftware





In other words, question your data assumptions and test for model robustness not just adequacy.
(2/n)
doi.org/10.1093/evol...

In other words, question your data assumptions and test for model robustness not just adequacy.
(2/n)
doi.org/10.1093/evol...
🔗 doi.org/10.1093/molbev/msaf184
#evobio #molbio

🔗 doi.org/10.1093/molbev/msaf184
#evobio #molbio
We wish you a delicious breakfast for tomorrow, while mulling over these lessons... 😉


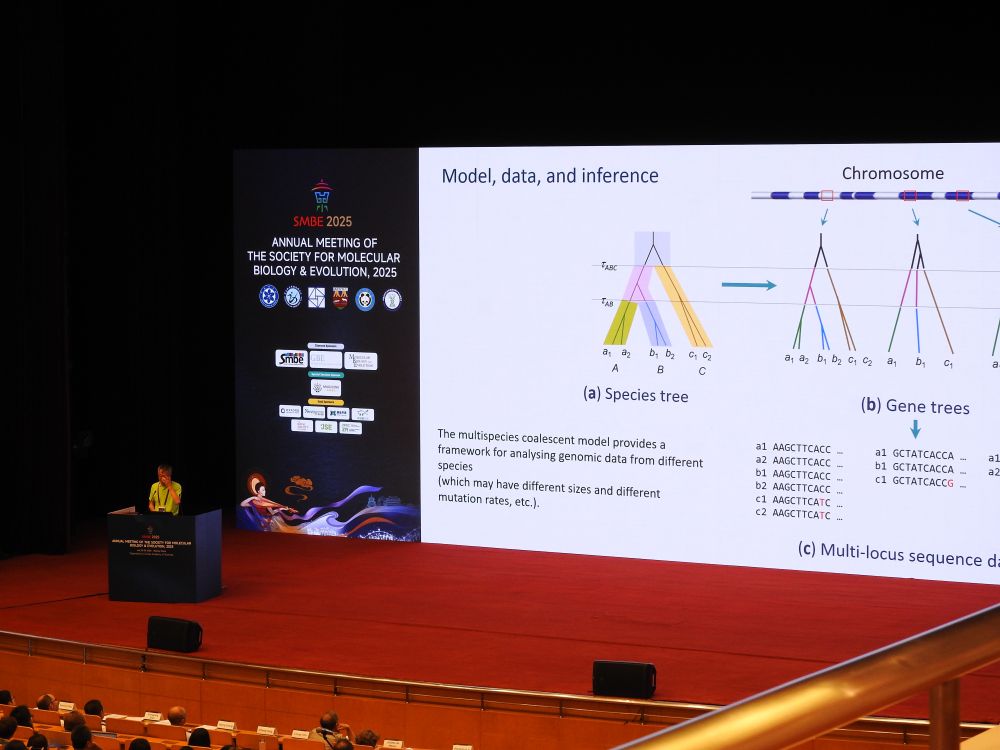

We wish you a delicious breakfast for tomorrow, while mulling over these lessons... 😉
🔗 doi.org/10.1093/molbev/msaf121
#evobio #molbio #geneflow
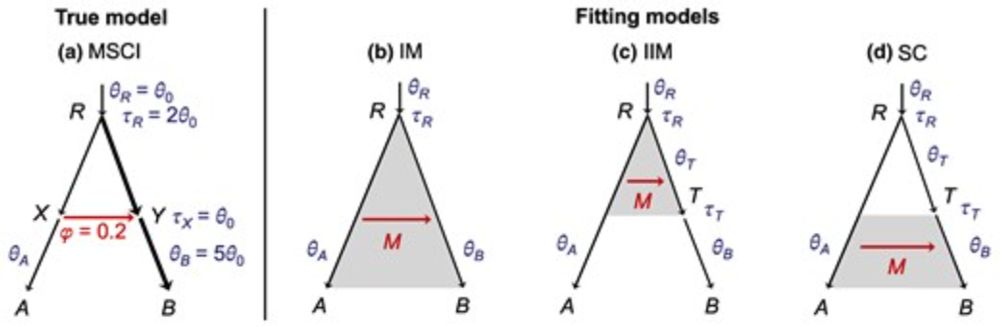
🔗 doi.org/10.1093/molbev/msaf121
#evobio #molbio #geneflow
"Six impossible things to believe before breakfast, about gene flow"
20.07.2025 | 18h00

"Six impossible things to believe before breakfast, about gene flow"
20.07.2025 | 18h00







Thank you to all the attendees, instructors (@anaserrasilva.bsky.social, @sabifo4.bsky.social, Tomas Flouri and Yuttapong Thawornwattana) and to our wonderful TA @lauramulvey.bsky.social!

Thank you to all the attendees, instructors (@anaserrasilva.bsky.social, @sabifo4.bsky.social, Tomas Flouri and Yuttapong Thawornwattana) and to our wonderful TA @lauramulvey.bsky.social!
We started with an introduction to the Multispecies Coalescent by Yuttapong Thawornwattana.






When: April 29-30, 2025
Where: University College London
There are limited places, so please pre-register by March 14th!!!
Find more details here: systass.org/workshop-bay...

You may want to check the TREES project led by the dos Reis Lab on integrating morphology and genomes for timetree inference! 🦴🧬💻Project supervisors: @mariodosreis.bsky.social & Ziheng Yang. More info in link 🔽
You may want to check the TREES project led by the dos Reis Lab on integrating morphology and genomes for timetree inference! 🦴🧬💻Project supervisors: @mariodosreis.bsky.social & Ziheng Yang. More info in link 🔽
Huge thanks to all participants for an inspiring [email protected]

Huge thanks to all participants for an inspiring [email protected]
Thank you to our incredible instructors for sharing their knowledge and making complex topics approachable and engaging! 🙌
And a big shoutout to all participants—Thanks for joining us this week🎉 @sabifo4.bsky.social @zihengyang.bsky.social

Thank you to our incredible instructors for sharing their knowledge and making complex topics approachable and engaging! 🙌
And a big shoutout to all participants—Thanks for joining us this week🎉 @sabifo4.bsky.social @zihengyang.bsky.social
Today, we’re honored to have one of the giants of Phylogenomics 🧬✨, @zihengyang.bsky.social, kicking off the day with two lectures on the Multispecies Coalescent Model.
Day3 will continue with an hands-on session led by Paschalia Kapli and Tomas Flouri🚀

Today, we’re honored to have one of the giants of Phylogenomics 🧬✨, @zihengyang.bsky.social, kicking off the day with two lectures on the Multispecies Coalescent Model.
Day3 will continue with an hands-on session led by Paschalia Kapli and Tomas Flouri🚀
Excited for the insights Day 3 will bring—stay tuned! 🚀

Excited for the insights Day 3 will bring—stay tuned! 🚀
The Phylogenomics course has just started :) @sabifo4.bsky.social @zihengyang.bsky.social


github.com/abacus-gene/...
github.com/abacus-gene/...
The Ziheng Yang Lab will be using this social account to post news about the software we develop, upcoming workshops, and general news in evolutionary biology and computational methods 💻🧬🦴
Keep an eye on this site, we will be posting some news soon!
The Ziheng Yang Lab will be using this social account to post news about the software we develop, upcoming workshops, and general news in evolutionary biology and computational methods 💻🧬🦴
Keep an eye on this site, we will be posting some news soon!

